| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,418,997 – 20,419,074 |
| Length | 77 |
| Max. P | 0.999506 |

| Location | 20,418,997 – 20,419,074 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 62.39 |
| Shannon entropy | 0.71811 |
| G+C content | 0.38888 |
| Mean single sequence MFE | -17.91 |
| Consensus MFE | -12.74 |
| Energy contribution | -10.94 |
| Covariance contribution | -1.80 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
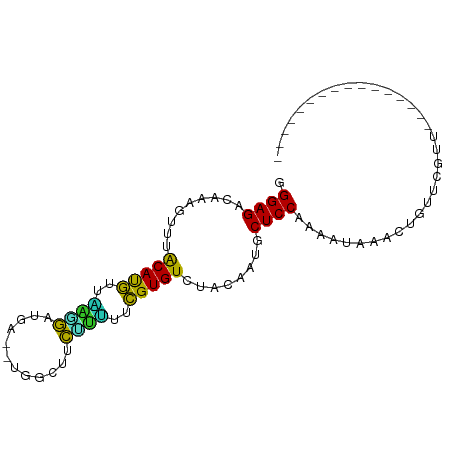
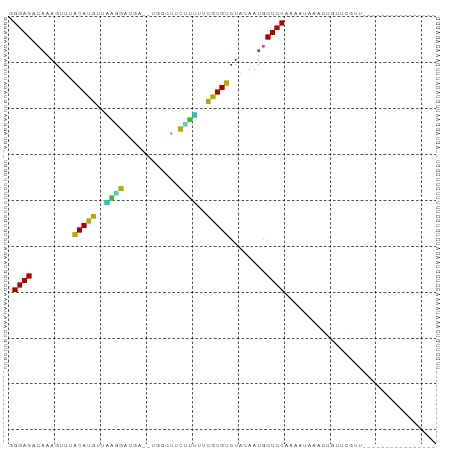
>dm3.chr2L 20418997 77 + 23011544 GGGAGACAAAGUUUACAUGUUGAAGAUGAUGUGGCUUCUUUUUCGUGUCUACAAUGCUCCAAAAUAAAUUGUUCGUU---------------- .((((.((..((..(((((..(((((.(......).)))))..)))))..))..)))))).................---------------- ( -18.10, z-score = -1.59, R) >droSim1.chr2L 19989032 77 + 22036055 GGGAGACAAAGUUUACAUGUUAAAGAUGAUGUUGCUUCUUUUUCGUGUCUACAAUGCUCCAAAAUAAACUGUUCGUU---------------- .((((.((..((..(((((..(((((.(......).)))))..)))))..))..)))))).................---------------- ( -17.70, z-score = -2.00, R) >droSec1.super_18 306631 77 + 1342155 GGGAGACAAAGUUUACACGUUAAAGAUGACGUGGCUUCUUUUUCGUGUCUACAAUGCUCCAAAAUAAACUGUUCGUU---------------- .((((.((..((..(((((..(((((.(......).)))))..)))))..))..)))))).................---------------- ( -19.50, z-score = -2.02, R) >droYak2.chr2R 6785202 74 + 21139217 GGGAGACAAAGUUUACAUGUUAAGGAUG---UGGCUUCUUUUUCGUGUCUACAAUGCUCCAACAUAAACUGUUCGUU---------------- .((((.((..((..(((((..(((((..---.....)))))..)))))..))..)))))).................---------------- ( -17.80, z-score = -1.57, R) >droEre2.scaffold_4845 18577147 72 - 22589142 CGGAGACAAAGUUUACAUGCUAAGGUCC-----ACAUCUUUUUCGUGUCUACAAUGCUCCAAGAAAAAUGUUUCGUU---------------- .((((.((..((..(((((..((((...-----....))))..)))))..))..))))))..((((....))))...---------------- ( -16.10, z-score = -1.94, R) >droAna3.scaffold_12916 3954039 75 + 16180835 UGGAGACAAAGUUUACAUGUUGAAGUCCA--UAGAAUCUUCCUCGUGUCUACAACGCUCCUAGAACAGCCUCCAACU---------------- (((((.....((..(((((..((((....--......))))..)))))..))...(((........))))))))...---------------- ( -17.90, z-score = -2.10, R) >dp4.chr4_group4 2666485 71 + 6586962 UGGAGCCAAAGUG-GCAUAUUUCUGAACC--AACCUUCAGAUUUGUGUCUCAAAUACUCCGAUAAUAACACGAU------------------- .((((.......(-(((((..((((((..--....))))))..)))))).......))))..............------------------- ( -19.94, z-score = -3.74, R) >droWil1.scaffold_180764 2080118 90 + 3949147 CGGAGCCAAAGUA-GCAUAUUUAGUUUUC--AUUCUUACAAUUUGUGUCUCAAAUGCUCCGAGAUGAACUUUAAAUCCGAACGGCAUCCUGUU .((((((...((.-....((((((..(((--((.(((...(((((.....))))).....))))))))..))))))....))))).))).... ( -16.10, z-score = -0.39, R) >droVir3.scaffold_12963 2581790 73 - 20206255 AGGAGUCAAAGUG-GCAUAUUUUGGGAAU--UUAAC-CGAAUUUGUGUCUCAAAUGCUCCAAAACUAAUUAAAAGAC---------------- .(((((....(.(-(((((.(((((....--....)-))))..)))))).)....))))).................---------------- ( -19.00, z-score = -3.03, R) >droMoj3.scaffold_6500 9688258 73 - 32352404 UGGAGUCAAAGUG-GCAUAUUCCGGGAUU--UCCUCUC-GAUCUGUGUCUCAAAUACUCCCAAAUAAACCCAAAGAC---------------- .(((((....(.(-(((((...(((((..--...))))-)...)))))).)....))))).................---------------- ( -17.00, z-score = -1.54, R) >consensus GGGAGACAAAGUUUACAUGUUAAGGAUGA__UGGCUUCUUUUUCGUGUCUACAAUGCUCCAAAAUAAACUGUUCGUU________________ .((((.........(((((..((((............))))..)))))........))))................................. (-12.74 = -10.94 + -1.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:13 2011