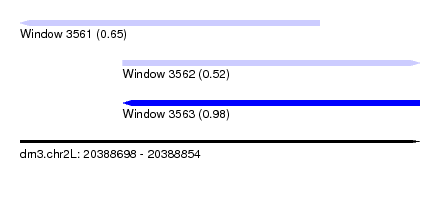
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,388,698 – 20,388,854 |
| Length | 156 |
| Max. P | 0.979999 |

| Location | 20,388,698 – 20,388,815 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.38 |
| Shannon entropy | 0.63561 |
| G+C content | 0.42462 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -7.68 |
| Energy contribution | -8.70 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20388698 117 - 23011544 UUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCAAACAGUGCGUAUUAUUAAUGCGAGUGCCCGUGGCUAAUAAUUAUUAGCCAUUACAAUCGGAUGCAUAAGUAAAAACACACAC ((((((......))))))....(((......))).....(((.((.(((((.((((((.((...(((((((((....)))))))))..)).))....)))).)))))..)).))).. ( -30.10, z-score = -2.11, R) >droAna3.scaffold_12916 8715677 109 + 16180835 --UUUAUGUGGCUUGUGGCACGG-CAACAAUCGAUUAUGUGACAGAUGAUUUGAGC-CCCGUUCGAAGCUAUUUAUCAAUCAGCAUUAUAGUGCCAUCCACAAGUACACAAGC---- --....((((((((((((...((-((.............(((.(((((.(((((((-...)))))))..))))).))).............))))..)))))))).))))...---- ( -31.77, z-score = -2.46, R) >droEre2.scaffold_4845 18552060 94 + 22589142 CUCUGAAACAGCUCAAAGCAAAGCCAGACACGGCGCACAGUGCGUAU----------------------UAAUAAUUAUUAGCCAUU-CAAUCGGAUGCAUAAGUAAAAACACAAAC .(((((....((.....))............(((((.....))....----------------------((((....)))))))...-...)))))..................... ( -12.20, z-score = 0.75, R) >droYak2.chr2R 6760192 101 - 21139217 ----------------UUCUGAAACAGCCACGGCACACAGUGCGUAUUAAUAAUGCGACUGCCCAUGGCUAAUAAUUAUUAGCCAUUUCAAUCGGAUGCAUAAGUAAAAACACAAAC ----------------.(((((....((....))...((((.(((((.....)))))))))...(((((((((....))))))))).....)))))..................... ( -24.50, z-score = -2.48, R) >droSec1.super_18 281817 117 - 1342155 UUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCACACAGUGCGUAUUAUUAAUGCGAGUGCCCGCGGCUAAUAAUUAUUAGCCAUUACAAUCGGAUGCAUAAGUAAAAACACACAC .(((((......)))))(((...((.((...(((((....(((((.......))))).))))).))(((((((....))))))).........)).))).................. ( -32.30, z-score = -2.86, R) >droSim1.chr2L 19962468 117 - 22036055 UUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCACACAGUGCGUAUUAUUAAUGCGAGUGCCCGCGGCUAAUAAUUAUUAGCCAUUACAAUCGGAUGCAUAAGUAAAAACACACAC .(((((......)))))(((...((.((...(((((....(((((.......))))).))))).))(((((((....))))))).........)).))).................. ( -32.30, z-score = -2.86, R) >consensus UUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCACACAGUGCGUAUUAUUAAUGCGAGUGCCCGCGGCUAAUAAUUAUUAGCCAUUACAAUCGGAUGCAUAAGUAAAAACACACAC ................................(((.....)))(((((......(((......)))(((((((....)))))))..........))))).................. ( -7.68 = -8.70 + 1.02)
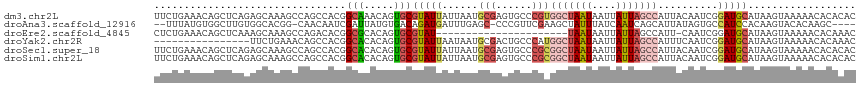
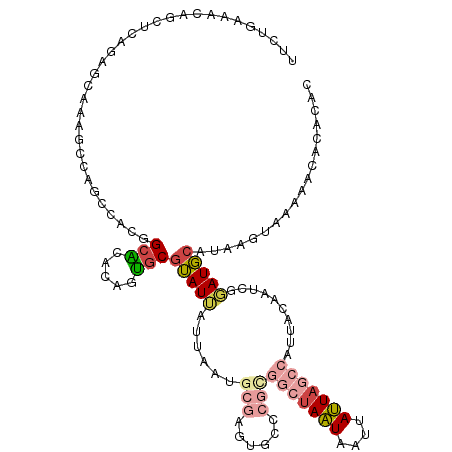
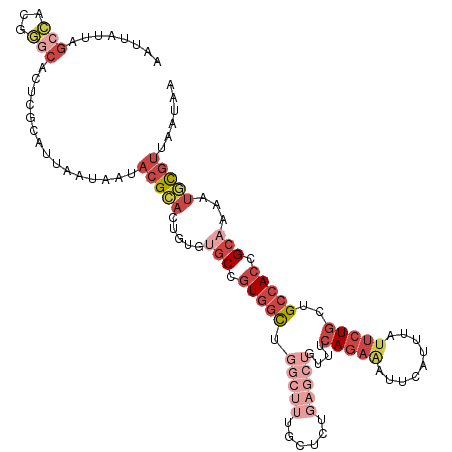
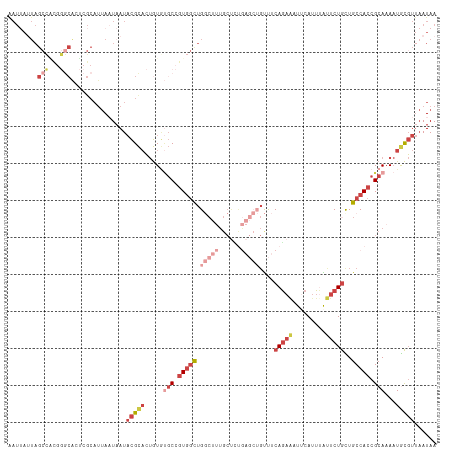
| Location | 20,388,738 – 20,388,854 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.45 |
| Shannon entropy | 0.59954 |
| G+C content | 0.42708 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -10.71 |
| Energy contribution | -13.55 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.523078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20388738 116 + 23011544 AAUUAUUAGCCACGGGCACUCGCAUUAAUAAUACGCACUGUUUGCCGUGGCUGGCUUUGCUCUGAGCUGUUUCAGAAUUCCAUUUAUUCUGCUGCCACCGCAAAAUGCGUUAAUAA .((((((((...(((....)))..))))))))(((((...(((((.(((((.(((((......)))))....((((((.......))))))..))))).))))).)))))...... ( -34.20, z-score = -2.08, R) >droAna3.scaffold_12916 8715713 109 - 16180835 GAUAAAUAGCUUCGAACGGG-GCUCAAAUCAUCUGUCACAUAAUCGAUUGUUG-CCGUGCCACAA---GCCACAUAAAUA-ACAGGCACUAUUGCCACCGCAAA-UAUUUCCAUAA (((....((((((....)))-)))...)))...............((...(((-(.(((.(....---).))).......-...((((....))))...)))).-....))..... ( -17.70, z-score = 0.51, R) >droEre2.scaffold_4845 18552099 94 - 22589142 ----------------------AAUUAUUAAUACGCACUGUGCGCCGUGUCUGGCUUUGCUUUGAGCUGUUUCAGAGUUUCAUUUAUUCUGUUGCCACCGCAAAAUGCGUUAAUAA ----------------------..((((((..(((((...((((..(((...(((((......)))))((..((((((.......))))))..)))))))))...))))))))))) ( -26.20, z-score = -2.27, R) >droYak2.chr2R 6760232 100 + 21139217 AAUUAUUAGCCAUGGGCAGUCGCAUUAUUAAUACGCACUGUGUGCCGUGGCUG----------------UUUCAGAAUUUCAUUAGUUCUGUUGCCACCGCAAAAUACGUUAAUAA ..((((((((.....(((((.((...........))))))).(((.(((((..----------------...(((((((.....)))))))..))))).)))......)))))))) ( -30.00, z-score = -2.75, R) >droSec1.super_18 281857 116 + 1342155 AAUUAUUAGCCGCGGGCACUCGCAUUAAUAAUACGCACUGUGUGCCGUGGCUGGCUUUGCUCUGAGCUGUUUCAGAAAUUCAUUUAUUCUGCUGCCACCGCAAAAUGCGUUAAUAA .((((((((..((((....)))).))))))))(((((.(...(((.(((((.(((((......)))))....(((((.........)))))..))))).))).).)))))...... ( -33.60, z-score = -1.13, R) >droSim1.chr2L 19962508 116 + 22036055 AAUUAUUAGCCGCGGGCACUCGCAUUAAUAAUACGCACUGUGUGCCGUGGCUGGCUUUGCUCUGAGCUGUUUCAGAAAUUCAUUUAUUCUGCUGCCACCGCAAAAUGCGUUAAUAA .((((((((..((((....)))).))))))))(((((.(...(((.(((((.(((((......)))))....(((((.........)))))..))))).))).).)))))...... ( -33.60, z-score = -1.13, R) >consensus AAUUAUUAGCCACGGGCACUCGCAUUAAUAAUACGCACUGUGUGCCGUGGCUGGCUUUGCUCUGAGCUGUUUCAGAAAUUCAUUUAUUCUGCUGCCACCGCAAAAUGCGUUAAUAA ........(((...)))...............(((((.....(((.(((((.(((((......)))))....(((((.........)))))..))))).)))...)))))...... (-10.71 = -13.55 + 2.84)

| Location | 20,388,738 – 20,388,854 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 69.45 |
| Shannon entropy | 0.59954 |
| G+C content | 0.42708 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -16.01 |
| Energy contribution | -17.20 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.979999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
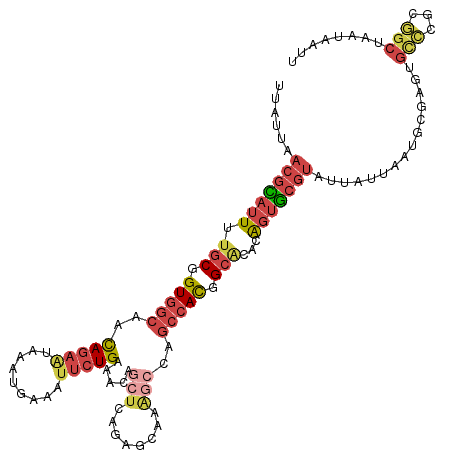
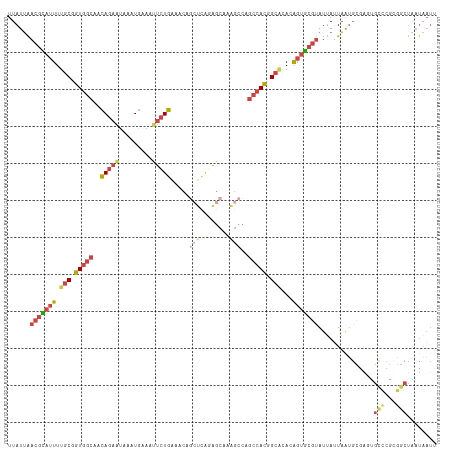
>dm3.chr2L 20388738 116 - 23011544 UUAUUAACGCAUUUUGCGGUGGCAGCAGAAUAAAUGGAAUUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCAAACAGUGCGUAUUAUUAAUGCGAGUGCCCGUGGCUAAUAAUU ((((((.(((.....))).((((..((((((.......)))))).....((.....))...))))(((((((((.....(((((.......)))))..)).))))))))))))).. ( -36.40, z-score = -2.18, R) >droAna3.scaffold_12916 8715713 109 + 16180835 UUAUGGAAAUA-UUUGCGGUGGCAAUAGUGCCUGU-UAUUUAUGUGGC---UUGUGGCACGG-CAACAAUCGAUUAUGUGACAGAUGAUUUGAGC-CCCGUUCGAAGCUAUUUAUC ..((((.....-...((((.(((....(((((.((-(((....)))))---....)))))(.-...)..((((((((.......)))).))))))-)))))......))))..... ( -26.64, z-score = 0.01, R) >droEre2.scaffold_4845 18552099 94 + 22589142 UUAUUAACGCAUUUUGCGGUGGCAACAGAAUAAAUGAAACUCUGAAACAGCUCAAAGCAAAGCCAGACACGGCGCACAGUGCGUAUUAAUAAUU---------------------- (((((((((((((.((((.((((..((((...........)))).....((.....))...)))......).)))).)))))))..))))))..---------------------- ( -23.70, z-score = -2.02, R) >droYak2.chr2R 6760232 100 - 21139217 UUAUUAACGUAUUUUGCGGUGGCAACAGAACUAAUGAAAUUCUGAAA----------------CAGCCACGGCACACAGUGCGUAUUAAUAAUGCGACUGCCCAUGGCUAAUAAUU ((((((.(((....(((.(((((..(((((.........)))))...----------------..))))).)))..((((.(((((.....)))))))))...)))..)))))).. ( -26.10, z-score = -1.94, R) >droSec1.super_18 281857 116 - 1342155 UUAUUAACGCAUUUUGCGGUGGCAGCAGAAUAAAUGAAUUUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCACACAGUGCGUAUUAUUAAUGCGAGUGCCCGCGGCUAAUAAUU ......(((((((.(((.(((((..(((((.........))))).....((.....)).......))))).)))...)))))))(((((((.((((......))))..))))))). ( -36.20, z-score = -2.10, R) >droSim1.chr2L 19962508 116 - 22036055 UUAUUAACGCAUUUUGCGGUGGCAGCAGAAUAAAUGAAUUUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCACACAGUGCGUAUUAUUAAUGCGAGUGCCCGCGGCUAAUAAUU ......(((((((.(((.(((((..(((((.........))))).....((.....)).......))))).)))...)))))))(((((((.((((......))))..))))))). ( -36.20, z-score = -2.10, R) >consensus UUAUUAACGCAUUUUGCGGUGGCAACAGAAUAAAUGAAAUUCUGAAACAGCUCAGAGCAAAGCCAGCCACGGCACACAGUGCGUAUUAUUAAUGCGAGUGCCCGCGGCUAAUAAUU ......(((((((.(((.(((((..(((((.........))))).....(((........)))..))))).)))...)))))))...............(((...)))........ (-16.01 = -17.20 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:11 2011