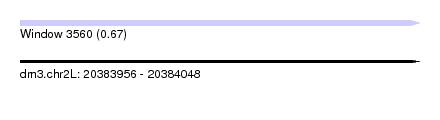
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,383,956 – 20,384,048 |
| Length | 92 |
| Max. P | 0.672081 |

| Location | 20,383,956 – 20,384,048 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.42 |
| Shannon entropy | 0.52813 |
| G+C content | 0.51650 |
| Mean single sequence MFE | -33.43 |
| Consensus MFE | -13.68 |
| Energy contribution | -14.56 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
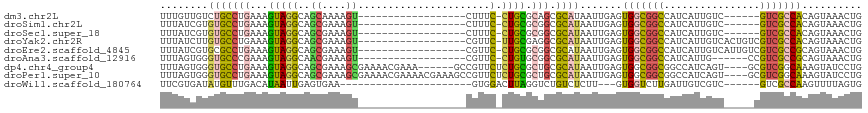
>dm3.chr2L 20383956 92 + 23011544 UUUGUUGUCUGCCUGAAAGUAGGCAGCAAAAGU------------------CUUUC-CUGCGCAGCGCAUAAUUGAGUGGCGGCCAUCAUUGUC------GUCGCCACAGUAAACUG ((((((((((((......))))))))))))..(------------------(..(.-.((((...))))..)..))((((((((((....))..------))))))))......... ( -31.70, z-score = -2.37, R) >droSim1.chr2L 19957762 92 + 22036055 UUUAUCGUGUGCCUGAAAGUAGGCAGCGAAAGU------------------CUUUC-CUGCGCGGCGCAUAAUUGAGUGGCGGCCAUCAUUGUC------GUCGCCACAGUAAACUG ......(((((((.(...(((((..((....))------------------....)-)))).))))))))......((((((((((....))..------))))))))......... ( -34.10, z-score = -2.56, R) >droSec1.super_18 277172 92 + 1342155 UUUAUCGUGUGCCUGAAAGUAGGCAGCGAAAGU------------------CUUUC-CUGCGCGGCGCAUAAUUGAGUGGCGGCCAUCAUUGUC------GUCGCCACAGUAAACUG ......(((((((.(...(((((..((....))------------------....)-)))).))))))))......((((((((((....))..------))))))))......... ( -34.10, z-score = -2.56, R) >droYak2.chr2R 6755483 98 + 21139217 UUUAUCUUGUGCCUGAAAGUAGGCAGCGAAAGU------------------CGUUC-UUGCGAGGCGCAUAAUUGAGUGGCGGCCAUCAUUGUCACUGUCGUCGCCACAGUAAACUG .......(((((((....(((((.((((.....------------------)))))-)))).))))))).......((((((((...((.......))..))))))))......... ( -33.20, z-score = -2.00, R) >droEre2.scaffold_4845 18547548 98 - 22589142 UUUAUCGUGCGCCUGAAAGUAGGCAGCGAAAGU------------------CGUUC-CUGCGCGGCGCAUAAUUGAGUGGCGGCCAUCAUUGUCAUUGUCGUCGCCGCAGUAAACUG ......(((((((.(...(((((.((((.....------------------)))))-)))).))))))))......((((((((...((.......))..))))))))......... ( -38.70, z-score = -2.66, R) >droAna3.scaffold_12916 8710874 92 - 16180835 UUUAGUGGGUGCCCGAAAGUAGGCAACGAAAGU------------------CGUUC-CUGUGCGGCGCAUAAUUGAGUGGCGGCCAUCAUUG------CCGUCGCCGCAGUAAACUG .(((((..(((((((...(((((.((((.....------------------)))))-)))).))).)))).)))))((((((((((....))------..))))))))......... ( -33.20, z-score = -1.41, R) >dp4.chr4_group4 2629645 107 + 6586962 UUUAGUGGGUGCCUGAAAGUAGGCAGCGAAAGCGAAAACGAAA------GCCGUUCUCUGCGCUGCGCAUAAUUGAGUGGCGGCGGCCAUCAGU----GCGUCGGCAAAGUAUCCUG ......((((((......(((((..((....))(((..(....------)...))))))))(((((((((...(((.((((....)))))))))----))).))))...)))))).. ( -39.40, z-score = -1.12, R) >droPer1.super_10 1639364 113 + 3432795 UUUAGUGGGUGCCUGAAAGUAGGCAGCGAAAGCGAAAACGAAAACGAAAGCCGUUCUCUGCGCUGCGCAUAAUUGAGUGGCGGCGGCCAUCAGU----GCGUCGGCAAAGUAUCCUG ......((((((..........(((((....))((.((((....(....).)))).)))))(((((((((...(((.((((....)))))))))----))).))))...)))))).. ( -39.40, z-score = -0.89, R) >droWil1.scaffold_180764 2031455 86 + 3949147 UUCGUGAUAUGUUUGACAUAAUUGAGUGAA----------------------GUGGACUUAGGUCUGUCUCUU---GUGGUCUUGAUUGUCGUC------GUCGCCAAGUUUUAGUG ...(((((......((((((((..((.((.----------------------.(((((....)))))..))..---.....))..))))).)))------)))))............ ( -17.10, z-score = -0.06, R) >consensus UUUAUCGUGUGCCUGAAAGUAGGCAGCGAAAGU__________________CGUUC_CUGCGCGGCGCAUAAUUGAGUGGCGGCCAUCAUUGUC_____CGUCGCCACAGUAAACUG .........((((((....))))))((....)).........................((((...))))........(((((((................))))))).......... (-13.68 = -14.56 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:09 2011