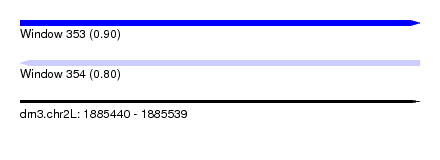
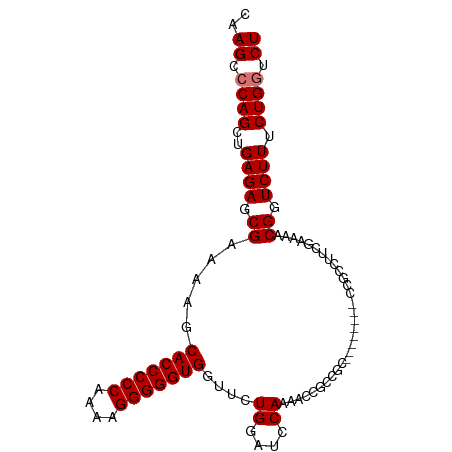
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,885,440 – 1,885,539 |
| Length | 99 |
| Max. P | 0.903355 |

| Location | 1,885,440 – 1,885,539 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.01 |
| Shannon entropy | 0.21888 |
| G+C content | 0.57774 |
| Mean single sequence MFE | -41.75 |
| Consensus MFE | -28.11 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1885440 99 + 23011544 AGACCAGAAAGACCGUUUUCGAAGGCAAACGGGCGGGCGGCGGAUUUGGAUCCAGAACCACCCCCUUUUGGGGGUGCUUUUCGCUCUCAGCUGGGCUUG ...((((..(((((((((.(....).))))))(((((.(((((.((((....)))).))((((((....))))))))).))))))))...))))..... ( -44.00, z-score = -2.44, R) >droEre2.scaffold_4929 1930966 84 + 26641161 AGACCAGAAAGAGCGUUUUCGAAGGCGG---------------AUUUGGAUCCAGAGCCACCCCCUUUUGGGGGUGGUUUUCGCUCUCAGCUGUGCUUG ((..(((..((((((...........((---------------((....))))((((((((((((....))))))))))))))))))...)))..)).. ( -38.40, z-score = -3.55, R) >droSec1.super_14 1833001 99 + 2068291 AGACCAGAAAGACCGUUUUCGAAGGCGGACGGGGGGGCGGCGGUUUUGGAUCCAGAACCACCCCCUUUUGGGGGUGCUUUUCGCUCUCAGCUGGGCUUG ...((((.....(((((..(....)..)))))((((((((.(((((((....)))))))((((((....)))))).....))))))))..))))..... ( -49.20, z-score = -3.16, R) >droSim1.chr2L_random 96872 91 + 909653 AGACCAGAAAGACCGUUUUCAAAGGCGG--------GCGGCGGUAUUGGAUCCAGAACCACCCCCUUUUGGGGGUGCUUUUCGCUCUCAGCUGGGCUUG ...((((..(((((((((....))))))--------((((.(((.(((....))).)))((((((....)))))).....)))))))...))))..... ( -35.40, z-score = -1.23, R) >consensus AGACCAGAAAGACCGUUUUCGAAGGCGG________GCGGCGGAUUUGGAUCCAGAACCACCCCCUUUUGGGGGUGCUUUUCGCUCUCAGCUGGGCUUG ...((((..(((.((.....((((((...............(((((((....)))))))((((((....)))))))))))))).)))...))))..... (-28.11 = -29.68 + 1.56)

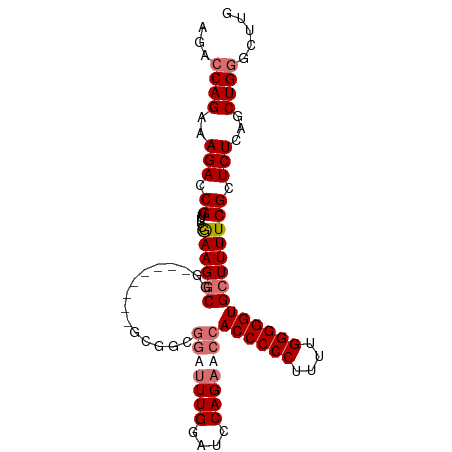
| Location | 1,885,440 – 1,885,539 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.01 |
| Shannon entropy | 0.21888 |
| G+C content | 0.57774 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -26.05 |
| Energy contribution | -26.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.795400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1885440 99 - 23011544 CAAGCCCAGCUGAGAGCGAAAAGCACCCCCAAAAGGGGGUGGUUCUGGAUCCAAAUCCGCCGCCCGCCCGUUUGCCUUCGAAAACGGUCUUUCUGGUCU ..((.((((..(((((((......((((((....))))))(((..(((((....)))))..))))))((((((........)))))))))).)))).)) ( -37.30, z-score = -1.93, R) >droEre2.scaffold_4929 1930966 84 - 26641161 CAAGCACAGCUGAGAGCGAAAACCACCCCCAAAAGGGGGUGGCUCUGGAUCCAAAU---------------CCGCCUUCGAAAACGCUCUUUCUGGUCU ...((.(((..(((((((....((((((((....))))))))....((((....))---------------))...........))))))).))))).. ( -35.40, z-score = -4.11, R) >droSec1.super_14 1833001 99 - 2068291 CAAGCCCAGCUGAGAGCGAAAAGCACCCCCAAAAGGGGGUGGUUCUGGAUCCAAAACCGCCGCCCCCCCGUCCGCCUUCGAAAACGGUCUUUCUGGUCU ..((.((((..(((((((....((((((((....))))))((((.((....)).)))))))))....((((.((....))...)))))))).)))).)) ( -32.90, z-score = -0.93, R) >droSim1.chr2L_random 96872 91 - 909653 CAAGCCCAGCUGAGAGCGAAAAGCACCCCCAAAAGGGGGUGGUUCUGGAUCCAAUACCGCCGC--------CCGCCUUUGAAAACGGUCUUUCUGGUCU ..((.((((..(((((((....((((((((....))))))(((..((....))..))))))))--------(((..........))))))).)))).)) ( -30.40, z-score = -0.78, R) >consensus CAAGCCCAGCUGAGAGCGAAAAGCACCCCCAAAAGGGGGUGGUUCUGGAUCCAAAACCGCCGC________CCGCCUUCGAAAACGGUCUUUCUGGUCU ..((.((((..((((.((.....(((((((....)))))))....((....))...............................)).)))).)))).)) (-26.05 = -26.30 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:34 2011