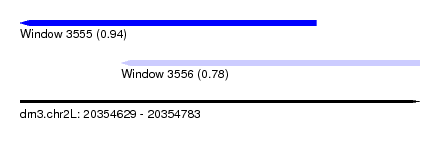
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,354,629 – 20,354,783 |
| Length | 154 |
| Max. P | 0.939958 |

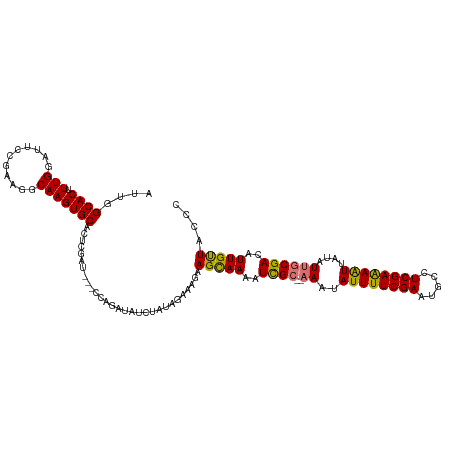
| Location | 20,354,629 – 20,354,743 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Shannon entropy | 0.28301 |
| G+C content | 0.31882 |
| Mean single sequence MFE | -17.83 |
| Consensus MFE | -13.99 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20354629 114 - 23011544 UCAGAUAUCUAUAGAAAGAAGUAAAAUCGC---AAUAUUUUCGAAUGCCUCGAAAAUUAUAUUACGACAUUGUUACUCACAA-AAACCCCCAAUAUUCCAAAGCUUAAUUGAAAUGGU .................((.((((..(((.---(((((((((((.....))))))...))))).))).....))))))....-..............(((...(......)...))). ( -11.90, z-score = 0.19, R) >droEre2.scaffold_4845 18519362 117 + 22589142 -CCAACAUUGCACGUAAAAAGCAAAAUUGCAAAAAUAUUUUCGAAAAACUCGAGAGUUAUAUUGCGACAUUGCUACCCACAAUCAAACCUAAAUAUUCCAAAGCAAAAUCCAAAUCUU -..................(((((..((((((....((((((((.....))))))))....))))))..)))))............................................ ( -18.60, z-score = -3.42, R) >droSec1.super_18 248246 115 - 1342155 CGAGAUAUCUAUAGAAAGAAGCAAAAUCGC--AAAUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAA-AAAACCUCAAUAUUCCAAAGAUAAAUCCAACUGGU .(((...(((......)))(((((..((((--((..((((((((.....))))))))....))))))..)))))........-.....)))..................((....)). ( -21.20, z-score = -3.11, R) >droSim1.chr2L 19928937 115 - 22036055 CGAGAUAUCUAUAGAAAGAAGCAAAAUCGC--AAAUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAA-AAAACAUUAAUAUUCCAAAGCUAAAUCGAAAUGGU (((....(((......)))(((((..((((--((..((((((((.....))))))))....))))))..)))))........-.........................)))....... ( -19.60, z-score = -1.94, R) >consensus CCAGAUAUCUAUAGAAAGAAGCAAAAUCGC__AAAUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCCACAA_AAAACCUCAAUAUUCCAAAGCUAAAUCCAAAUGGU ...................(((((..((((......((((((((.....))))))))......))))..)))))............................................ (-13.99 = -13.30 + -0.69)

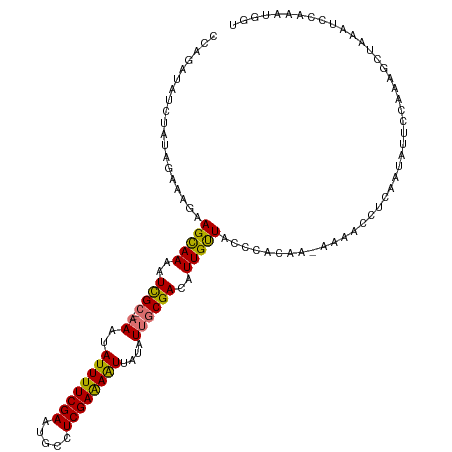
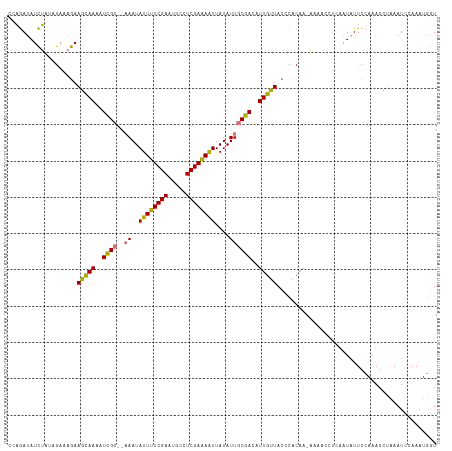
| Location | 20,354,668 – 20,354,783 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.98 |
| Shannon entropy | 0.25231 |
| G+C content | 0.37160 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -19.49 |
| Energy contribution | -18.80 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20354668 115 - 23011544 AUUGGCACAUUGGAUUCUAAAGGCAAGUGCACUCUAUACAUCAGAUAUCUAUAGAAAGAAGUAAAAUCGC---AAUAUUUUCGAAUGCCUCGAAAAUUAUAUUACGACAUUGUUACUC ....((((.(((...........)))))))..((((((.((.....)).))))))....(((((..(((.---(((((((((((.....))))))...))))).))).....))))). ( -18.10, z-score = -0.10, R) >droEre2.scaffold_4845 18519402 114 + 22589142 AUGGGCACUUUGGAUUCUGAAAGCAAGUGCACUAGA----UCCAACAUUGCACGUAAAAAGCAAAAUUGCAAAAAUAUUUUCGAAAAACUCGAGAGUUAUAUUGCGACAUUGCUACCC ..(((((..((((((.(((...((....))..))))----)))))...)))........(((((..((((((....((((((((.....))))))))....))))))..))))).)). ( -31.50, z-score = -3.50, R) >droSec1.super_18 248285 113 - 1342155 AUUGGCACUUUGGAUUCCGAAGGCAAGUGCACUCGAU---CGAGAUAUCUAUAGAAAGAAGCAAAAUCGC--AAAUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCC ....((.((((((...)))))))).......(((...---.)))...(((......)))(((((..((((--((..((((((((.....))))))))....))))))..))))).... ( -28.10, z-score = -2.00, R) >droSim1.chr2L 19928976 113 - 22036055 AUUGGCACUUUGGAUUCCGAAGGCAAGUGCACUCGAU---CGAGAUAUCUAUAGAAAGAAGCAAAAUCGC--AAAUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCC ....((.((((((...)))))))).......(((...---.)))...(((......)))(((((..((((--((..((((((((.....))))))))....))))))..))))).... ( -28.10, z-score = -2.00, R) >consensus AUUGGCACUUUGGAUUCCGAAGGCAAGUGCACUCGAU___CCAGAUAUCUAUAGAAAGAAGCAAAAUCGC__AAAUAUUUUCGAAUGCCUCGAAAAUUAUAUUGCGACAUUGUUACCC ....((((.(((...........))))))).............................(((((..((((......((((((((.....))))))))......))))..))))).... (-19.49 = -18.80 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:05 2011