
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,343,794 – 20,343,929 |
| Length | 135 |
| Max. P | 0.641425 |

| Location | 20,343,794 – 20,343,889 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.69 |
| Shannon entropy | 0.44819 |
| G+C content | 0.64125 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.69 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
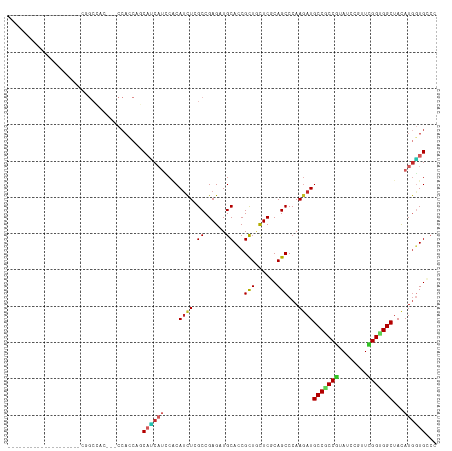
>dm3.chr2L 20343794 95 + 23011544 --------------------CGGCCAC---CCACCAGCAUCAUCCACAUUUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUACAUGGUGCCC --------------------.(((.((---(.....(((((.((..(.....)..)))))))...((((....)))).......(((((((........))))))).....)))))). ( -32.50, z-score = -1.87, R) >droSim1.chr2L 19918151 95 + 22036055 --------------------CGGCCAC---CCACCAGCAUCAUCCACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUACAUGGUGCCC --------------------.(((.((---(.......(((.....((((((...))))))....((((....))))....)))(((((((........))))))).....)))))). ( -33.30, z-score = -1.97, R) >droSec1.super_18 237408 95 + 1342155 --------------------CGGCCAC---CCACCAGCAUCAUCCACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUACAUGGUGCCC --------------------.(((.((---(.......(((.....((((((...))))))....((((....))))....)))(((((((........))))))).....)))))). ( -33.30, z-score = -1.97, R) >droYak2.chr2R 6715610 95 + 21139217 --------------------CGGCCAC---CCACCAGCACCAUCCACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCGUUCGGUGGCUACAUGGUGCCC --------------------.((....---...)).(((((((...((((((...))))))....((((....)))).......(((((((........)))))))...))))))).. ( -34.80, z-score = -2.13, R) >droEre2.scaffold_4845 18508493 95 - 22589142 --------------------CGGCCAC---CCACCAGCAUCAUCCGCAUCUCGCCGAGAUGCACCGCUGCUCCCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUAUAUGGUGCCC --------------------.(((.((---(.......(((....(((((((...)))))))...((((....))))....)))(((((((........))))))).....)))))). ( -36.10, z-score = -3.03, R) >droAna3.scaffold_12916 8667625 98 - 16180835 --------------------CGGCCACCCUCCACCACCAGCAUCCGCACCUGGCCGAGAUGCAUCGCUACUCGCAGCCCAAGAUGCCACCGUAUCCGAUUGGUGGCUACAUGGUGGCC --------------------.(((((((.....((((((((((((((.....).)).)))))((((((......)))....(((((....))))).)))))))))......))))))) ( -37.50, z-score = -2.23, R) >droPer1.super_10 1598487 116 + 3432795 CGGCCACACACCAGCACCACCAGCAGCA--CCAGCCUCACCAUGCCCAUCUAGCCGAGCUGCACCGCUGCUCGCAGCCCAAGAUGCCACCGUAUCCGUUUGGGGGCUACAUGGUGCCC .(((.........((.......))....--...))).(((((((......(((((((((.((...)).))))....((((((((((....)))))...)))))))))))))))))... ( -35.79, z-score = 0.19, R) >dp4.chr4_group4 2589152 116 + 6586962 CGGCCACACACCAGCACCACCAGCAGCA--CCAGCCUCACCAUGCCCAUCUAGCCGAGCUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCGUUUGGGGGCUACAUGGUGCCC .(((.........((.......))....--...))).(((((((......(((((((((.((...)).))))....((((((((((....)))))...)))))))))))))))))... ( -35.79, z-score = 0.64, R) >droVir3.scaffold_12963 8361226 86 - 20206255 -------------------------------CAGCCGCCGC-CGCACAUCUCGCCGAGCUGCAUCGUUAUUCCCAGCCCAAAAUGCCGCCAUAUCCCAUUGGCGGCUACAUAUUGGGC -------------------------------..((.((((.-((.......)).)).)).)).............((((((...(((((((........)))))))......)))))) ( -29.50, z-score = -1.93, R) >droGri2.scaffold_15126 3001559 80 - 8399593 -------------------------------------CUGC-CGCGCAUCUCGCCGAGCUGCAUCGCUACUCACAGCCCAAGAUGCCGCCAUAUCCGAUCGGUGGCUAUAUGCUGGCC -------------------------------------..((-((.((((.(((((((((.((((((((......)))....))))).)).........)))))))....)))))))). ( -26.10, z-score = -0.38, R) >consensus ____________________CGGCCAC___CCACCAGCAUCAUCCACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCGUUCGGUGGCUACAUGGUGCCC .....................................((((((....((((.((......))...(((......)))...))))(((((((........)))))))...))))))... (-18.39 = -18.69 + 0.30)


| Location | 20,343,815 – 20,343,929 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.73 |
| Shannon entropy | 0.56754 |
| G+C content | 0.61553 |
| Mean single sequence MFE | -44.29 |
| Consensus MFE | -20.96 |
| Energy contribution | -21.04 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
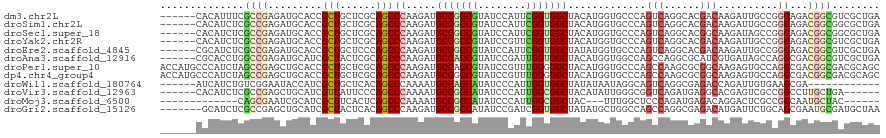
>dm3.chr2L 20343815 114 + 23011544 ------CACAUUUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUACAUGGUGCCCAGUCAGGCACGACAAGAUUGCCGGGAGACGGCGUCGCUGA ------........(((..((((((((((((....)))).......(((((((........))))))).....)))))...))).)))....((.(((.((((.....)))))))..)). ( -44.40, z-score = -0.95, R) >droSim1.chr2L 19918172 114 + 22036055 ------CACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUACAUGGUGCCCAGUCAGGCACGACAAGAUUGCCGGGAGACGGCGGCGCUGA ------..((((((...))))))....((((....))))...((.(((((((((.(((...(((..(((...(.(((((......))))).)..)).)..)))))).))))))))))).. ( -49.00, z-score = -1.61, R) >droSec1.super_18 237429 114 + 1342155 ------CACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUACAUGGUGCCCAGUCAGGCACGGCAAGAUAGCCGGGAGACGGCGGCGCUGA ------..((((((...))))))((.((((((.((...(((.....(((((((........)))))))......(((((......)))))(((......))))))...)))))))).)). ( -51.40, z-score = -1.81, R) >droYak2.chr2R 6715631 114 + 21139217 ------CACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCGUUCGGUGGCUACAUGGUGCCCAGUCAGGCACGACAAGAUUGCCGGGAGACGGCGUCGCUGA ------........(((..((((((((((((....)))).......(((((((........))))))).....)))))...))).)))....((.(((.((((.....)))))))..)). ( -44.40, z-score = -0.36, R) >droEre2.scaffold_4845 18508514 114 - 22589142 ------CGCAUCUCGCCGAGAUGCACCGCUGCUCCCAGCCCAAGAUGCCGCCGUAUCCAUUCGGUGGCUAUAUGGUGCCCAGUCAGGCACGACAAGAUUGCCGGGAGACGGCGUCGCUGA ------.(((((((...)))))))..((((((((((.((.((....(((((((........)))))))....))(((((......))))).........)).))))).)))))....... ( -51.80, z-score = -2.86, R) >droAna3.scaffold_12916 8667649 114 - 16180835 ------CGCACCUGGCCGAGAUGCAUCGCUACUCGCAGCCCAAGAUGCCACCGUAUCCGAUUGGUGGCUACAUGGUGGCCAGCCAGGCGCAUCGUGAUAGCCAGGCGACGGCGUCGCUGA ------.((.((((((((((..((...))..))))..((((.....(((((((........)))))))......).)))..)))))).))...(((((.(((.(....)))))))))... ( -47.80, z-score = -0.74, R) >droPer1.super_10 1598523 120 + 3432795 ACCAUGCCCAUCUAGCCGAGCUGCACCGCUGCUCGCAGCCCAAGAUGCCACCGUAUCCGUUUGGGGGCUACAUGGUGCCCAGCCAAGCGCGGCAAGAGUGCCAGGCGACGGCGACGCAGC ....(((((((.(((((((((.((...)).))))....((((((((((....)))))...)))))))))).))))((((..(((..((((.......))))..)))...))))..))).. ( -48.90, z-score = -0.24, R) >dp4.chr4_group4 2589188 120 + 6586962 ACCAUGCCCAUCUAGCCGAGCUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCGUUUGGGGGCUACAUGGUGCCCAGCCAAGCGCGGCAAGAGUGCCAGGCGACGGCGACGCAGC ....(((.(((((.(((((((.((...)).)))))..))...))))).((((((..((((((((((((........))))..))))))..((((....)))).))..))))))..))).. ( -50.10, z-score = -0.11, R) >droWil1.scaffold_180764 1980401 102 + 3949147 ------AUCAUCUGUCGGAAUACCAUCGCUGCUCACAGCCCAAAAUGCCACCAUAUCCCAUUGGUGGCUAUAUAAUAGGCAGUCAGGCGAGACCAGAUUGUGAAGCGA------------ ------..........((....)).(((((..((((((((......(((((((........))))))).........))).(((......))).....))))))))))------------ ( -30.26, z-score = -1.40, R) >droVir3.scaffold_12963 8361238 108 - 20206255 ------CACAUCUCGCCGAGCUGCAUCGUUAUUCCCAGCCCAAAAUGCCGCCAUAUCCCAUUGGCGGCUACAUAUUGGGCGGUCAGAUGAGGCACGAGUCGCCGGCCUUGCUGA------ ------..(((((.(((((......))..........((((((...(((((((........)))))))......))))))))).)))))((((.((......))))))......------ ( -42.10, z-score = -2.08, R) >droMoj3.scaffold_6500 11807591 98 - 32352404 -------------CAGCGAAUCGCAUCGCUUCACUCAGCCCAAAAUGCCGCCAUAUCCCAUUGGCGGCUAC---UUUGGCUCCCAGAUGAGACAGGACUCGCCGGCAAUGCUAC------ -------------.........((((.(((......((((......(((((((........)))))))...---...))))....(.((((......)))).)))).))))...------ ( -30.30, z-score = -1.35, R) >droGri2.scaffold_15126 3001566 113 - 8399593 -------GCAUCUCGCCGAGCUGCAUCGCUACUCACAGCCCAAGAUGCCGCCAUAUCCGAUCGGUGGCUAUAUGCUGGCCAGCCAGGCGAGACAUGAUUCUGCAGCGAAUGCGAUGCUAA -------(((..((((...(((((((((...(((.((((.......((((((..........)))))).....))))(((.....))))))...)))...))))))....)))))))... ( -41.00, z-score = -0.83, R) >consensus ______CACAUCUCGCCGAGAUGCACCGCUGCUCGCAGCCCAAGAUGCCGCCGUAUCCGUUCGGUGGCUACAUGGUGCCCAGUCAGGCGCGACAAGAUUGCCGGGCGACGGCGACGCUGA ..............((((.........(((......)))((.....(((((((........))))))).............(((......)))..........))...))))........ (-20.96 = -21.04 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:04 2011