
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,341,659 – 20,341,776 |
| Length | 117 |
| Max. P | 0.626075 |

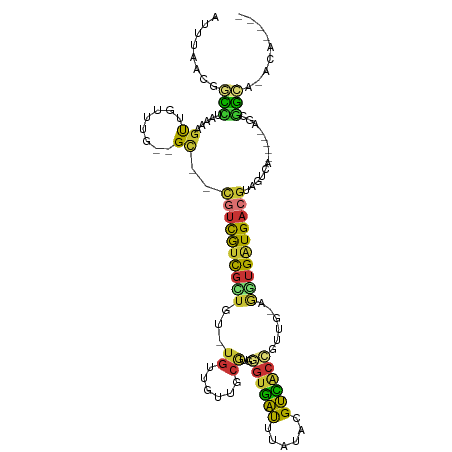
| Location | 20,341,659 – 20,341,758 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.06 |
| Shannon entropy | 0.65295 |
| G+C content | 0.47925 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -11.49 |
| Energy contribution | -10.91 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20341659 99 - 23011544 AUUUAACGGCCUAAAAGUUGUUUG--GC--CGUCGUCGCUGU-UGUUGUUGCGUGGGUGAUUUAUACGUCACCGUUG-AGGUGAUGACGUCGUCA-----AGCGGCA-ACA---- .....((((((............)--))--))).........-(((((((((...(..(((.....(((((((....-.)))))))..)))..).-----.))))))-)))---- ( -35.70, z-score = -2.20, R) >droSim1.chr2L 19915988 99 - 22036055 AUUUAACGGCCUAAAAGUUGUUUG--GC--CGUCGUCGCUGU-UGUUGUUGCGUGGGUGAUUUAUACGUCACCGUUG-GGGUGAUGACGUCGUCA-----AGCGGCA-ACA---- .....((((((............)--))--))).........-(((((((((...(..(((.....(((((((....-.)))))))..)))..).-----.))))))-)))---- ( -35.10, z-score = -1.77, R) >droSec1.super_18 235252 99 - 1342155 AUUUAACGGCCUAAAAGCUGUUUG--GC--CGUCGUCGCUGU-UGUUGUUGCGUGGGUGAUUUAUACGUCACCGUUG-GGGUGAUGACGUCGUCA-----AGCGGCA-ACA---- ................((((((((--((--((((((((((..-.((....))...((((((......))))))....-.))))))))))..))))-----)))))).-...---- ( -37.50, z-score = -2.25, R) >droYak2.chr2R 6713478 98 - 21139217 AUUUAACGGCCUAAAAGUGGUUU---GC--CGUUGUCGCUAU-UGUUGUUGCGUGGGUGAUUUAUACGUCACCGUUG-AGGUGAUGACGUAGGCU-----AGCGGCC-ACA---- .......((((......((((((---((--.((..(((((..-.((....))...((((((......))))))....-.)))))..)))))))))-----)..))))-...---- ( -32.00, z-score = -1.14, R) >droEre2.scaffold_4845 18506451 104 + 22589142 AUUUAACGGCCUAAAAGUGGUUUG--GC--CGUCGGUGGUGU-UGUUGCUGCGUGGGUGAUUUAUACGUCACCGUUG-AGGUGAUGACGUAGACGUAGACAGCGGCA-ACA---- ..(..((((((............)--))--)))..)...(((-(((((((((..............(((((((....-.)))))))(((....))).).))))))))-)))---- ( -39.10, z-score = -2.47, R) >droAna3.scaffold_12916 8665398 101 + 16180835 AUUUAAC-ACACAAAUGUUUGGUG--GU----UUGUUGUUGU-UAUUGUUGCUUUAGUGAUUUAUACGUCACUCCUG-AAGUGGUGACGUAGUCAGUGCAGGCGGUG-ACA---- ....(((-(.((((((........--))----))))))))((-(((((((((...(.((((...((((((((..(..-..)..)))))))))))).)))).))))))-)).---- ( -30.10, z-score = -1.77, R) >droMoj3.scaffold_6500 11804973 107 + 32352404 AUUUAAC-AUUUAAAGACCCCG----GCUACGCUGUUUGUGUAUGUUACGUCACGAGUUGUGUGUGAGCGGCGGCGGCUAUUGAUGACGU-CGUGCGUAAGAGAGCGCACAAU-- ......(-(((...((.(((((----.((((((.....)))))..(((((.(((.....))))))))..).))).))))...))))....-.((((((......))))))...-- ( -31.20, z-score = 0.39, R) >droGri2.scaffold_15126 2999237 110 + 8399593 AUUUAAC-AUUUAAAAGCGUCAAGCCGCUAUAUUAUUGCCGU-UGUUACGUCAUUAGUUGCG-AUGGGCA--GGCGGCAUUUGAUGACGUAUGCAUUUUAAGGGACACACACACA .......-.(((((((((((((.((((((.......((((((-(((.((.......)).)))-)).))))--))))))......)))))).....)))))))............. ( -26.11, z-score = -0.34, R) >consensus AUUUAACGGCCUAAAAGUUGUUUG__GC__CGUCGUCGCUGU_UGUUGUUGCGUGGGUGAUUUAUACGUCACCGUUG_AGGUGAUGACGUAGUCA_____AGCGGCA_ACA____ ..............................((((((((((...((......))..((((((......))))))......)))))))))).......................... (-11.49 = -10.91 + -0.58)


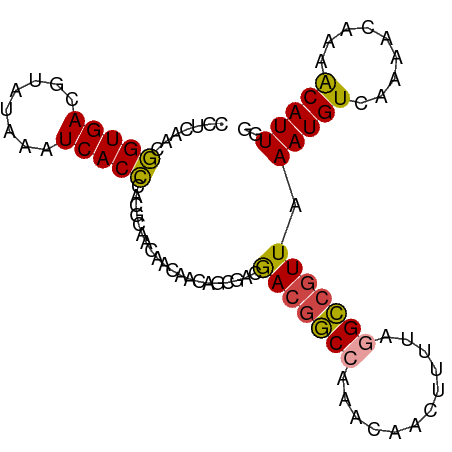
| Location | 20,341,683 – 20,341,776 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 86.81 |
| Shannon entropy | 0.25155 |
| G+C content | 0.43441 |
| Mean single sequence MFE | -18.09 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.02 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20341683 93 + 23011544 CCUCAACGGUGACGUAUAAAUCACCCACGCAACAACAACAGCGACGACGGCCAAACAACUUUUAGGCCGUUAAAUGUCAAAACAAAACAUUGG .......(.((((((............(((..........)))..(((((((............)))))))..))))))...).......... ( -19.80, z-score = -1.94, R) >droSim1.chr2L 19916012 93 + 22036055 CCCCAACGGUGACGUAUAAAUCACCCACGCAACAACAACAGCGACGACGGCCAAACAACUUUUAGGCCGUUAAAUGUCAAAACAAAACAUUGG ..((((.(((((........)))))..(((..........)))..(((((((............)))))))..................)))) ( -21.10, z-score = -2.24, R) >droSec1.super_18 235276 93 + 1342155 CCCCAACGGUGACGUAUAAAUCACCCACGCAACAACAACAGCGACGACGGCCAAACAGCUUUUAGGCCGUUAAAUGUCAAAACAAAACAUUGG ..((((.(((((........)))))..(((..........)))..(((((((............)))))))..................)))) ( -21.10, z-score = -1.94, R) >droYak2.chr2R 6713502 92 + 21139217 CCUCAACGGUGACGUAUAAAUCACCCACGCAACAACAAUAGCGACAACGGC-AAACCACUUUUAGGCCGUUAAAUGUGAAAACAAAACAUUGG .....(((....))).....((((...(((..........)))..((((((-.............))))))....)))).............. ( -18.12, z-score = -1.29, R) >droEre2.scaffold_4845 18506480 93 - 22589142 CCUCAACGGUGACGUAUAAAUCACCCACGCAGCAACAACACCACCGACGGCCAAACCACUUUUAGGCCGUUAAAUGUCAAAACAAAACAUUGA ..((((.(((((........)))))....................(((((((............)))))))..................)))) ( -17.50, z-score = -1.36, R) >droAna3.scaffold_12916 8665427 90 - 16180835 CUUCAGGAGUGACGUAUAAAUCACUAAAGCAACAAUAACAACAACAA--ACCACCAAACAUUUGUGU-GUUAAAUGUCAAAACAAAGCAUUUA .......(((((........)))))...((.....(((((...((((--(..........))))).)-))))..(((....)))..))..... ( -10.90, z-score = 0.04, R) >consensus CCUCAACGGUGACGUAUAAAUCACCCACGCAACAACAACAGCGACGACGGCCAAACAACUUUUAGGCCGUUAAAUGUCAAAACAAAACAUUGG .......(((((........)))))....................(((((((............))))))).(((((.........))))).. (-14.39 = -14.02 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:02 2011