
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,327,754 – 20,327,865 |
| Length | 111 |
| Max. P | 0.547657 |

| Location | 20,327,754 – 20,327,865 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.47 |
| Shannon entropy | 0.64123 |
| G+C content | 0.50113 |
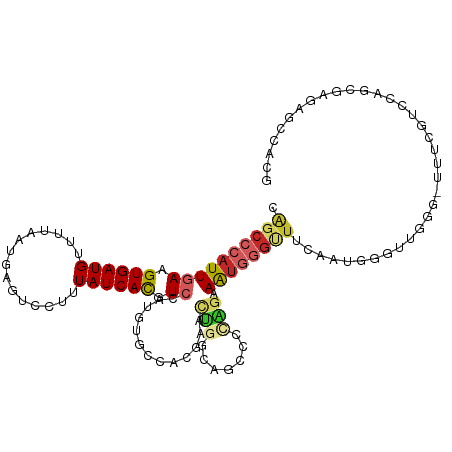
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -9.43 |
| Energy contribution | -9.98 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
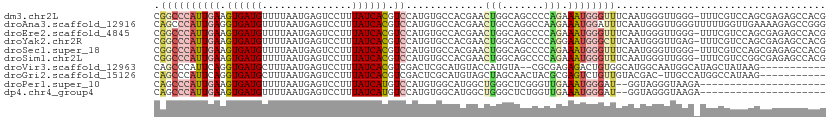
>dm3.chr2L 20327754 111 - 23011544 CGGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGGCAGCCCCAGAAAUGGGUUUCAAUGGGUUGGG-UUUCGUCCAGCGAGAGCCACG .(((((((((((((((((...............)))))).......(((((.....)))))..((((....)))).)))))))))))((.-(((((.....))))).))... ( -42.36, z-score = -3.08, R) >droAna3.scaffold_12916 8650875 112 + 16180835 CAGCCCAUUGAGGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGCCAGGCCAAGAAAUGGAUUUCAAUGGGUUGGGUUUUUGGUUGAAAAGAGCCGGG ((((((((((((((((((...............))))))((((((..(((...........)))......))))))))))))))))))(((((((.......)))))))... ( -37.16, z-score = -1.89, R) >droEre2.scaffold_4845 18492259 111 + 22589142 CGGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGGCAGCCCCAGAAAUGGGUUUCAAUGGGUUGGG-UUUCGUCCAGCGAGAGCCACG .(((((((((((((((((...............)))))).......(((((.....)))))..((((....)))).)))))))))))((.-(((((.....))))).))... ( -42.36, z-score = -3.08, R) >droYak2.chr2R 6699502 111 - 21139217 CGGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGGCAGCCCCAGGAAUGGGCUUCAAUGGGUUGAG-UUUCGUCCAGCGAGAGCCACG ((((((((((((((((((...............))))))((((((.(((((.....))))).((...)).))))))))))))))))))..-(((((.....)))))...... ( -38.86, z-score = -2.05, R) >droSec1.super_18 217037 111 - 1342155 CGGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGGCAGCCCCAGAAAUGGGUUUCAAUGGGUUGGG-UUUCGUCCAGCGAGAGCCACG .(((((((((((((((((...............)))))).......(((((.....)))))..((((....)))).)))))))))))((.-(((((.....))))).))... ( -42.36, z-score = -3.08, R) >droSim1.chr2L 19900565 111 - 22036055 CGGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGGCAGCCCCAGAAAUGGGUUUCAAUGGGUUGGG-UUUCGUCCGGCGAGAGCCACG ((((((((((((((((((...............)))))).......(((((.....)))))..((((....)))).))))))))))))((-......))(((....)))... ( -43.66, z-score = -3.01, R) >droVir3.scaffold_12963 8332043 99 + 20206255 CAGCCCAUUCAGGUGAUGCUUUAAUGAGUCCUUUAUCACGUCGACUCGCAUGUACCAUGUA--CGCGAGAGACUGUGGCAUGGCAAUGGCAUAGCUAUAAG----------- ..(((((.....(((((((((....)))).....)))))(((..(((((..((((...)))--)))))).))))).)))(((((.........)))))...----------- ( -27.00, z-score = -0.30, R) >droGri2.scaffold_15126 2980549 100 + 8399593 CAGCCCAUUCAGGUGAUGCUUUAAUGAGUCCUUUAUCACGUCGACUCGCAUGUAGCUAGCAACUACGCGAGUCUGUUGUACGAC-UUGCCAUGGCCAUAAG----------- ..(((....(((((..(((..((.(((........)))....(((((((..((((.......)))))))))))))..)))..))-)))....)))......----------- ( -25.40, z-score = -0.43, R) >droPer1.super_10 1581921 89 - 3432795 CAGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCAUGUCCAUGUGGCAUGGCUGGGCUCGGGUUGAAAUGGGAU--GGUAGGGUAAGA--------------------- (((((((((((((.....)))))))((((((....(((((((.....)))))))..)))))))))))).........--............--------------------- ( -27.90, z-score = -1.46, R) >dp4.chr4_group4 2572366 89 - 6586962 CAGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCAUGUCCAUGUGGCAUGGCUGGGCUCUGGUUGAAAUGGGAU--GGUAGGGUAAGA--------------------- ..((((..((..((..(((((((((((((((....(((((((.....)))))))..))))))..)))))))))..))--..))))))....--------------------- ( -26.30, z-score = -0.95, R) >consensus CAGCCCAUUGAAGUGAUGUUUUAAUGAGUCCUUUAUCACGUCCAUGUGCCACGAACUGGCAGCCCCAGAAAUGGGUUUCAAUGGGUUGGG_UUUCGUCCAGCGAGAGCCACG .((((((((((.((((((...............)))))).)).............(((.......))).))))))))................................... ( -9.43 = -9.98 + 0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:54:00 2011