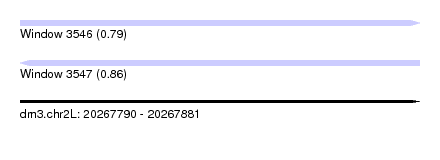
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,267,790 – 20,267,881 |
| Length | 91 |
| Max. P | 0.863546 |

| Location | 20,267,790 – 20,267,881 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Shannon entropy | 0.39194 |
| G+C content | 0.51747 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.80 |
| Energy contribution | -21.76 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
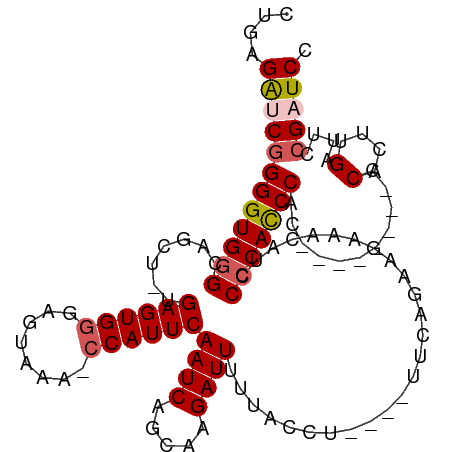
>dm3.chr2L 20267790 91 + 23011544 CUGAGAUCGGGGUGGGCAGCU-UGAGUGGGACUAAAACCAUUCAAUCGGCAAGAUUUUUACCU----UUC------------CCCACCCAC--------CCACUUGAUUCCGAUCC ....((((((((((((..(.(-(((((((........)))))))).)((.(((........))----).)------------)...)))))--------).........)))))). ( -30.90, z-score = -2.32, R) >droSim1.chr2L 19859154 102 + 22036055 CUGAGAUCGGGGUGGGCCGCU-UGAGUGGGAGUAAA-CCAUUCAAUCAGCAAGAUUUUUACCU----UUCAGAAGAAACAUCCCCACCCAC--------CCACUUGAUUCCGAUCC ....((((((((((((..(((-(((((((.......-)))))))...)))............(----(((....))))....)))))))..--------.(....)....))))). ( -31.10, z-score = -1.82, R) >droSec1.super_18 176026 103 + 1342155 CUGAGAUCGGGGUGGGCCCCGGUGAGUGGGAGUAAA-CCAUUCAAUCAGCAAGAUUUUUACCU----UUCAGAAGAAACAUCCCCACCCAC--------CCACUUGAUUCCGAUCC ....((((((((((((....(((((((((.......-))))))((((.....))))...)))(----(((....))))....)))))))..--------.(....)....))))). ( -30.40, z-score = -0.96, R) >droYak2.chr2R 6658320 114 + 21139217 CUGAGGCCGGGGUGGGCAGCU-UGAGUGGAAGUAAA-CCAUUCAAUCAGCAAGAUUUUUACCUUUACCCCCACAUUCACAUUCACACCCACACCCAACCCCACCAGCACCCGAUCU .((.((..((((((((..(((-(((((((.......-)))))))...)))(((........)))......................))))).)))..)).)).............. ( -26.10, z-score = -0.79, R) >droEre2.scaffold_4845 18444601 100 - 22589142 CUGAGA-CGGGGUGGGCAGCU-UGAGUGCAAGUAAA-CCAUUCAAUCAGCAAGAUUUUUACCUG---CUCCUCAUCCCCAUCCCCAUCC----------CCACCUGUUCAGGAUCC (((((.-.((((((((.....-((((.(((.(((((-......((((.....))))))))).))---)..))))........)))))))----------)......)))))..... ( -28.02, z-score = -1.03, R) >consensus CUGAGAUCGGGGUGGGCAGCU_UGAGUGGGAGUAAA_CCAUUCAAUCAGCAAGAUUUUUACCU____UUCAGAAGAAACAUCCCCACCCAC________CCACUUGAUUCCGAUCC ....((((((((((((..(((.(((((((........)))))))...))).((........))...................)))))))...........(....)....))))). (-20.80 = -21.76 + 0.96)


| Location | 20,267,790 – 20,267,881 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.11 |
| Shannon entropy | 0.39194 |
| G+C content | 0.51747 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -18.34 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
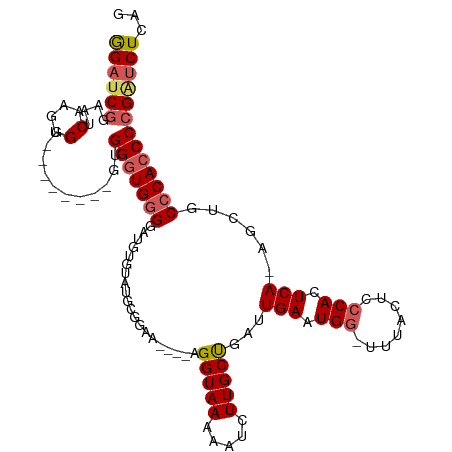
>dm3.chr2L 20267790 91 - 23011544 GGAUCGGAAUCAAGUGG--------GUGGGUGGG------------GAA----AGGUAAAAAUCUUGCCGAUUGAAUGGUUUUAGUCCCACUCA-AGCUGCCCACCCCGAUCUCAG (((((((......((((--------(((((((((------------(.(----((((....))))).))(((((((....))))))))))))).-....)))))).)))))))... ( -34.60, z-score = -2.35, R) >droSim1.chr2L 19859154 102 - 22036055 GGAUCGGAAUCAAGUGG--------GUGGGUGGGGAUGUUUCUUCUGAA----AGGUAAAAAUCUUGCUGAUUGAAUGG-UUUACUCCCACUCA-AGCGGCCCACCCCGAUCUCAG (((((((......((((--------(((((((((..((..(((((....----.(((((.....)))))....))).))-..))..))))))).-....)))))).)))))))... ( -36.00, z-score = -1.81, R) >droSec1.super_18 176026 103 - 1342155 GGAUCGGAAUCAAGUGG--------GUGGGUGGGGAUGUUUCUUCUGAA----AGGUAAAAAUCUUGCUGAUUGAAUGG-UUUACUCCCACUCACCGGGGCCCACCCCGAUCUCAG (((((...........(--------(((((((((..((..(((((....----.(((((.....)))))....))).))-..))..))))))))))((((....)))))))))... ( -40.20, z-score = -2.20, R) >droYak2.chr2R 6658320 114 - 21139217 AGAUCGGGUGCUGGUGGGGUUGGGUGUGGGUGUGAAUGUGAAUGUGGGGGUAAAGGUAAAAAUCUUGCUGAUUGAAUGG-UUUACUUCCACUCA-AGCUGCCCACCCCGGCCUCAG ..(((((...)))))(((((((((.((((((((.....(((..(((((((((((......((((.....))))......-))))))))))))))-.)).))))))))))))))).. ( -46.70, z-score = -3.25, R) >droEre2.scaffold_4845 18444601 100 + 22589142 GGAUCCUGAACAGGUGG----------GGAUGGGGAUGGGGAUGAGGAG---CAGGUAAAAAUCUUGCUGAUUGAAUGG-UUUACUUGCACUCA-AGCUGCCCACCCCG-UCUCAG ....(((....)))..(----------((((((((.((((.(((((..(---((((((((((((.....))))......-))))))))).))))-...).)))))))))-)))).. ( -40.70, z-score = -2.72, R) >consensus GGAUCGGAAUCAAGUGG________GUGGGUGGGGAUGUGUAUGCGGAA____AGGUAAAAAUCUUGCUGAUUGAAUGG_UUUACUCCCACUCA_AGCUGCCCACCCCGAUCUCAG ((((((....(....)...........(((((((....................(((((.....)))))...(((.(((........))).)))......)))))))))))))... (-18.34 = -19.10 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:58 2011