| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,238,933 – 20,239,036 |
| Length | 103 |
| Max. P | 0.623736 |

| Location | 20,238,933 – 20,239,036 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 63.58 |
| Shannon entropy | 0.70667 |
| G+C content | 0.47839 |
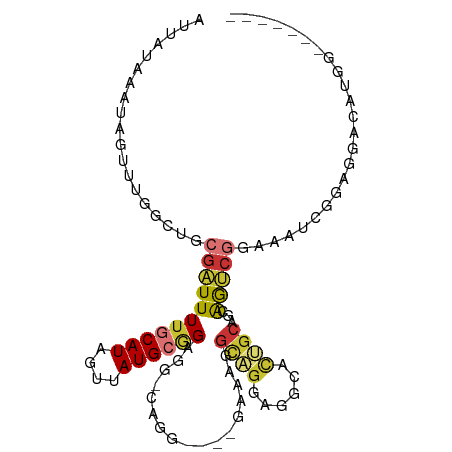
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -7.45 |
| Energy contribution | -7.51 |
| Covariance contribution | 0.06 |
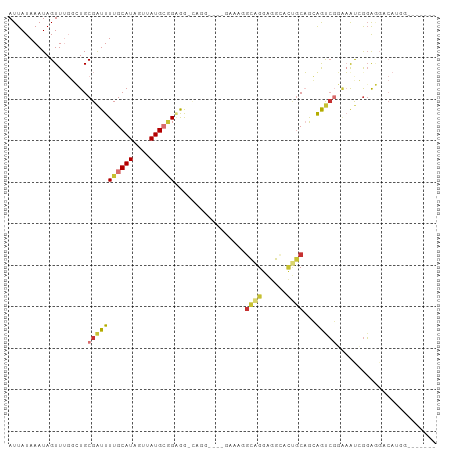
| Combinations/Pair | 1.47 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
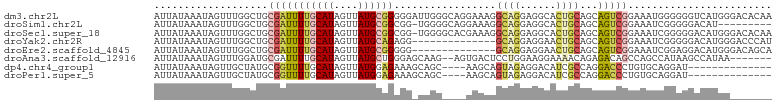
>dm3.chr2L 20238933 103 + 23011544 AUUAUAAAUAGUUUGGCUGCGAUUUUGCAUAGUUAUGCGGGGGAUUGGGCAGGAAAGGCAGGAGGCACUGCAGCAGUCGGAAAUGGGGGGUCAUGGGACACAA ...........(((((((((..((((((.(((((........))))).))))))...((((......)))).)))))))))........(((....))).... ( -27.70, z-score = -2.46, R) >droSim1.chr2L 19828597 93 + 22036055 AUUAUAAAUAGUUUGGCUGCGAUUUUGCAUAGUUAUGCGGCGG-UGGGGCAGGAAAGGCAGGAGGCACUGCAGCAGUCGGAAAUCGGGGGACAU--------- ...........(((((((((.....(((((....)))))((((-((..((.......))......)))))).))))))))).....(....)..--------- ( -26.10, z-score = -2.02, R) >droSec1.super_18 144011 102 + 1342155 AUUAUAAAUAGUUUGGCUGCGAUUUUGCAUAGUUAUGCGGCGG-UGGGGCACGAAAGGCAGGAGGCACUGCAGCAGUCGGAAAUCGGGGGACAUGGGACACAA ...........(((((((((.....(((((....)))))((((-((..((.(....)))......)))))).)))))))))..((((....).)))....... ( -29.80, z-score = -2.45, R) >droYak2.chr2R 6628063 89 + 21139217 AUUAUAAAUAGUUUGGCUGCGAUUUUGCAUAGUUAUGCAGAGG--------------GCAGGAGGAACUGCAGCAGUCGGAAAUCGGGGGACAUGGGACCCAU ...........(((((((((..((((((((....)))))))).--------------((((......)))).))))))))).....(((..(...)..))).. ( -30.80, z-score = -3.88, R) >droEre2.scaffold_4845 18413352 89 - 22589142 AUUAUAAAUAGUUUGGCUGCGAUUUUGCAUAGUUAUGCGGGGG--------------GCAGGAGGAACUGCAGCAGUCGGAAAUCGGAGGACAUGGGACAGCA ...........(((((((((..((((((((....)))))))).--------------((((......)))).)))))))))...................... ( -23.90, z-score = -2.32, R) >droAna3.scaffold_12916 7510790 94 - 16180835 AUUAUAAAUAGUUUGGAUGCGAUUUUGCAUAGUUAUGCUGGGAGCAAG--AGUGACUCCUGGAAGGAAAACAGAGACAGCCAGCCAUAAGCCAUAA------- .............((((((((....)))))..((((((((((((((..--..)).)))(((.........)))......)))).))))).)))...------- ( -18.90, z-score = 0.08, R) >dp4.chr4_group1 2257948 85 + 5278887 AUUAUAAAUAGUUGCUAUGCGGUUUUGCAUAGUUAUGGAGAAAGCAGC----AAGCAGUAGAGGACAUCGCCAGGACCCUGUGCAGGAU-------------- ...........(((((.(((..((((.(((....))).)))).)))..----.)))))...........(((((....))).)).....-------------- ( -18.10, z-score = 0.48, R) >droPer1.super_5 3822323 85 - 6813705 AUUAUAAAUAGUUGCUAUGCGGUUUUGCAUAGUUAUGGAGAAAGCAGC----AAGCAGUAGAGGACAUCGCCAGGACCCUGUGCAGGAU-------------- ...........(((((.(((..((((.(((....))).)))).)))..----.)))))...........(((((....))).)).....-------------- ( -18.10, z-score = 0.48, R) >consensus AUUAUAAAUAGUUUGGCUGCGAUUUUGCAUAGUUAUGCGGAGG_CAGG____GAAAGGCAGGAGGCACUGCAGCAGUCGGAAAUCGGAGGACAUGG_______ ...................(((((((((((....)))))).................((((......))))...)))))........................ ( -7.45 = -7.51 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:50 2011