
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,130,221 – 20,130,317 |
| Length | 96 |
| Max. P | 0.711322 |

| Location | 20,130,221 – 20,130,317 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 70.44 |
| Shannon entropy | 0.58160 |
| G+C content | 0.36796 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -7.47 |
| Energy contribution | -7.05 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
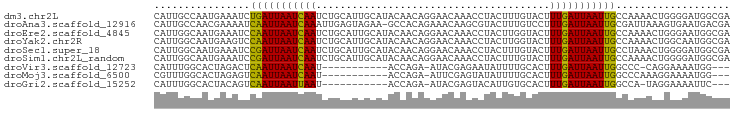
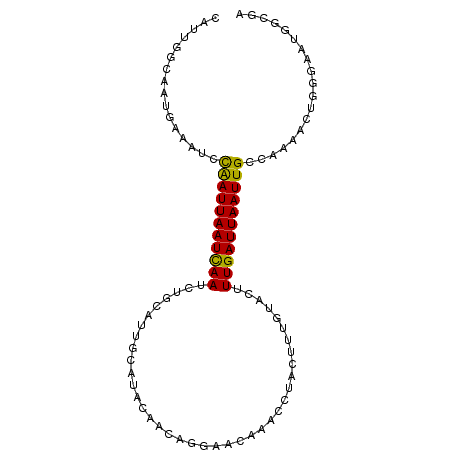
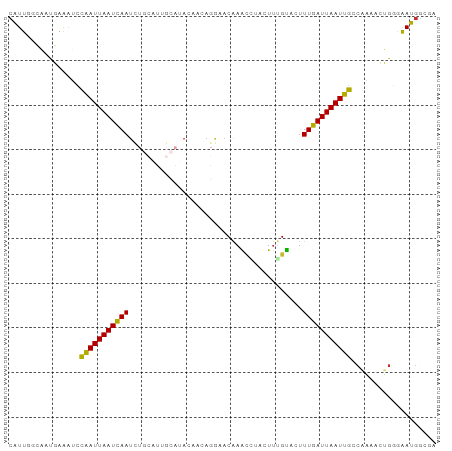
>dm3.chr2L 20130221 96 - 23011544 CAUUGCCAAUGAAAUCUGAUUAAUCAAUCUGCAUUGCAUACAACAGGAACAAACCUACUUUGUACUUUGAUUAAUUGCCAAAACUGGGGAUGGCGA ..((((((.........((((((((((..(((((((....))).(((......)))....))))..)))))))))).(((....)))...)))))) ( -21.80, z-score = -2.07, R) >droAna3.scaffold_12916 7478540 95 + 16180835 CAUUGCCAACGAAAAUCAAUUAAUCAAAUUGAGUAGAA-GCCACAGAAACAAGCGUACUUUGUCCUUUGAUUAAUUGCGAUUAAAGUGAAUGACGA ((((.....((.....((((((((((((..(((((...-((...........)).))))).....)))))))))))))).....))))........ ( -14.10, z-score = -0.12, R) >droEre2.scaffold_4845 18379021 96 + 22589142 CAUUGGCAAUGAAAUCCAAUUAAUCAAUCUGCAUUGCAUACAACAGGAACAAACCUACUUGGUACUUUGAUUAAUUGCCAAAACUGGGAAUGGCGA ..(((.((........(((((((((((..(((.(((....))).(((......))).....)))..)))))))))))(((....)))...)).))) ( -18.50, z-score = -0.72, R) >droYak2.chr2R 6592962 96 - 21139217 CAUUGGCAAUGAAGUCCAAUUAAUCAAUCUGCAUUGCAUACAACAGGAACAAACCUACUUGGUACUUUGAUUAAUUGCCAAAACUGGCAAUGGCGA (((((.((........(((((((((((..(((.(((....))).(((......))).....)))..))))))))))).......)).))))).... ( -20.86, z-score = -1.04, R) >droSec1.super_18 64323 96 - 1342155 CAUUGGCAAUGAAAUCCGAUUAAUCAAUCUGCAUUGCAUACAACAGGAACAAACCUACUUUGUACUUUGAUUAAUUGCCUAAACUGGGGAUGGCGA .........((..((((((((((((((..(((((((....))).(((......)))....))))..)))))))))).((......))))))..)). ( -20.40, z-score = -1.45, R) >droSim1.chr2L_random 790625 96 - 909653 CAUUGGCAAUGAAAUCCGAUUAAUCAAUCUGCAUUGCAUACAACAGGAACAAACCUACUUUGUACUUUGAUUAAUUGCCAAAACUGGGGAUGGCGA .........((..((((((((((((((..(((((((....))).(((......)))....))))..)))))))))).(((....)))))))..)). ( -20.80, z-score = -1.47, R) >droVir3.scaffold_12723 5765354 80 - 5802038 CAUUUGGCACUAGACUCAAUUAAUCAAU-----------ACCAGA-AUACGAGAAUAUUUUGCACUUUGAUUAAUUGGCCC-CAGGAAAAUGG--- (((((....((.(..((((((((((((.-----------..((((-(((......)))))))....))))))))))))..)-.))..))))).--- ( -18.80, z-score = -3.03, R) >droMoj3.scaffold_6500 11386036 81 - 32352404 CGUUUGGCACUAGAGUCAAUUAAUCAAU-----------ACCAGA-AUUCGAGUAUAUUUUGCACUUUGAUUAAUUGGCCCAAAGGAAAAUGG--- (.(((((.......(((((((((((((.-----------..((((-((........))))))....)))))))))))))))))).).......--- ( -20.91, z-score = -3.01, R) >droGri2.scaffold_15252 15435608 80 + 17193109 CAUUUGGCACUACAGUCAAUUAAUUAAU-----------ACCAGA-AUACGAGUACAUUGUGCACUUUGAUUAAUUGGCCA-UAGGAAAAUUC--- .........(((..(((((((((((((.-----------...((.-.(((((.....)))))..)))))))))))))))..-)))........--- ( -16.00, z-score = -1.88, R) >consensus CAUUGGCAAUGAAAUCCAAUUAAUCAAUCUGCAUUGCAUACAACAGGAACAAACCUACUUUGUACUUUGAUUAAUUGCCAAAACUGGGAAUGGCGA ................(((((((((((.......................................)))))))))))................... ( -7.47 = -7.05 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:44 2011