| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,075,041 – 20,075,174 |
| Length | 133 |
| Max. P | 0.986850 |

| Location | 20,075,041 – 20,075,174 |
|---|---|
| Length | 133 |
| Sequences | 11 |
| Columns | 153 |
| Reading direction | reverse |
| Mean pairwise identity | 55.73 |
| Shannon entropy | 0.94449 |
| G+C content | 0.45406 |
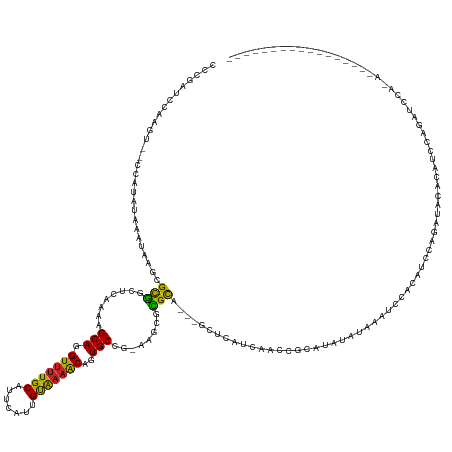
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -12.42 |
| Energy contribution | -11.44 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
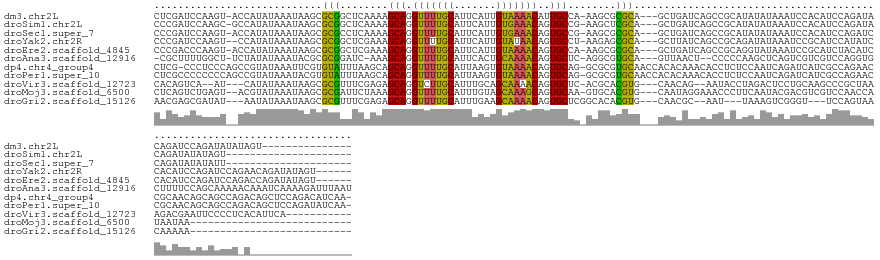
>dm3.chr2L 20075041 133 - 23011544 CUCGAUCCAAGU-ACCAUAUAAAUAAGCGCGGCUCAAAAGCAGGUUUUGCAUUCAUUGUAAAACAUUGCCA-AAGCGCGCA---GCUGAUCAGCCGCAUAUAUAAAUCCACAUCCAGAUACAGAUCCAGAUAUAUAGU--------------- ((.((((.....-...............((((((....(((..((((((((.....))))))))..((((.-....).)))---)))....))))))........(((........)))...)))).)).........--------------- ( -29.70, z-score = -2.12, R) >droSim1.chr2L 19730737 127 - 22036055 CCCGAUCCAAGC-GCCAUAUAAAUAAGCGCGGCUCAAAAGCAGGUUUUGCAUUCAUUGUGAAACAGUGCCG-AAGCUCGCA---GCUGAUCAGCCGCAUAUAUAAAUCCACAUCCAGAUACAGAUAUAUAGU--------------------- ..........((-.............))((((((....(((..((((..((.....))..))))..(((..-......)))---)))....))))))((((((..(((........)))....))))))...--------------------- ( -28.02, z-score = -0.67, R) >droSec1.super_7 3687698 127 - 3727775 CCCGAUCCAAGU-ACCAUAUAAAUAAGCGCGCCUCAAAAGCAGGUUUUGCAUUCAUUGUGAAACAGUGCCG-AAGCGCGCA---GCUGAUCAGCCGCAUAUAUAAAUCCACAUCCAGAUCCAGAUAUAUAUU--------------------- ...((((.....-.............((((((.((....(((.((((..((.....))..))))..))).)-).)))))).---(((....)))......................))))............--------------------- ( -29.80, z-score = -2.45, R) >droYak2.chr2R 6539207 141 - 21139217 CCCGAUCCAAGU--CCAUAUAAAUAAGCGCGGCUCGAAAGCAGGUUUUGCAUUCAUUGUAUAACAGUGCCU-AAGAGCGCA---GCUUAUCAGCCGCAGAUAUAAAUCCGCAUCCAUAUCCACAUCCAGAUCCAGAACAGAUAUAGU------ ...((((.....--..............((((((.((((((..(((.((((.....)))).))).((((..-....)))).---)))).)))))))).(((((............)))))........))))...............------ ( -31.20, z-score = -2.23, R) >droEre2.scaffold_4845 18333527 142 + 22589142 CCCGACCCAAGU-ACCAUAUAAAUAAGCGCGGCUCGAAAGCAGGUUUUGCAUUCAUUGUAAAACAGUGCCA-AAGCGCGCA---GCUGAUCAGCCGCAGGUAUAAAUCCGCAUCUACAUCCACAUCCAGAUCCAGACCAGAUAUAGU------ ............-....((((.....((((((((.((.(((..((((((((.....)))))))).((((..-....)))).---)))..)))))))).((.......)))).(((..(((........)))..))).....))))..------ ( -34.70, z-score = -2.33, R) >droAna3.scaffold_12916 7444431 144 + 16180835 -CGCUUUUGGCU-UCUAUAUAAAUACGCGCGGAUC-AAAGCAGGUUUUGCAUUCACUGCAAAACAGUGCUC-AGGCGUGCA---GUUAACU--CCCCCAAGCUCAGUCGUCGUCCAGGUGCUUUUCCAGCAAAAACAAAUCAAAAGAUUUAAU -..(((((((((-(..........(((((((..((-..((((.((((((((.....))))))))..)))).-.))))))).---)).....--.....)))).........((.....((((.....))))...))....))))))....... ( -31.65, z-score = -0.05, R) >dp4.chr4_group4 3000293 150 + 6586962 CUCG-CCCUCCCAGCCGUAUAAAUUCGUGUAUUUAAGCAGCAGGUUUUGCAUUAAGUGUAAAACAGUGCAG-GCGCGUGCAACCACACAAACACCUCUCCAAUCAGAUCAUCGCCAGAACCGCAACAGCAGCCAGACAGCUCCAGACAUCAA- ...(-(..((...(((((.(((((......))))).)).(((.(((((((((...)))))))))..))).)-))((.(((...............(((......)))...((....))...)))...)).....))..))............- ( -28.00, z-score = 0.26, R) >droPer1.super_10 817098 151 + 3432795 CUCGCCCCCCCCAGCCGUAUAAAUACGUGUAUUUAAGCAGCAGGUUUUGCAUUAAGUGUAAAACAGUGCAG-GCGCGUGCAACCACACAAACACCUCUCCAAUCAGAUCAUCGCCAGAACCGCAACAGCAGCCAGACAGCUCCAGAUAUCAA- ..((((..........((.((((((....)))))).)).(((.(((((((((...)))))))))..))).)-))).(((......))).................(((.(((((.......))....(.(((......))).).))))))..- ( -29.70, z-score = -0.65, R) >droVir3.scaffold_12723 4607156 131 - 5802038 CACAGUCA--AU---CAUAUAAAUAAGCGCGUUUCGAGAGCAGGUCUUGCAUUUGCAGCAAAACAGUGCUC-ACGCACGUG---CAACAG--AAUACCUAGACUCCUGCAAGCCCGCUAAAGACGAAUUCCCCUCACAUUCA----------- ....(((.--..---...........((((((..((.(((((.((.((((.......)))).))..)))))-.)).)))))---).....--...............((......))....)))((((.........)))).----------- ( -28.30, z-score = -0.78, R) >droMoj3.scaffold_6500 10574763 120 - 32352404 CUCAGUCUGAGU--ACGUAUAAAUAAGCGCGAUUCUAAAGCAGGUUUUGCAUUUGUAGCAAAGCAGUGCAA-GUGCACGUG---CAAUAGGAAACCCUUCAAUACGACGUCGUCCAACCAUAAUAA--------------------------- ....(..(((((--.(((((......((((.............(((((((.......))))))).((((..-..)))))))---)...(((....)))...))))))).)))..)...........--------------------------- ( -28.30, z-score = -1.04, R) >droGri2.scaffold_15126 3561995 112 - 8399593 AACGAGCGAUAU---AAUAUAAAUAAGCGCGUUUCGAGAGCAGGUUUUGCAUUUGAAGCAAAACAGUGCUCGGCACACGUG---CAACGC--AAU---UAAAGUCGGGU---UCCAGUAACAAAAA--------------------------- ...((((.....---...........((((((.....(((((.(((((((.......)))))))..))))).....)))))---)..(((--...---....).)).))---))............--------------------------- ( -29.20, z-score = -1.59, R) >consensus CCCGAUCCAAGU__CCAUAUAAAUAAGCGCGGCUCAAAAGCAGGUUUUGCAUUCAUUGUAAAACAGUGCCG_AAGCGCGCA___GCUCAUCAACCGCAUAUAUAAAUCCACAUCCAGAUACACAUCCAGAUCCA_A_________________ ............................(((........(((.(((((((.......)))))))..)))........)))......................................................................... (-12.42 = -11.44 + -0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:42 2011