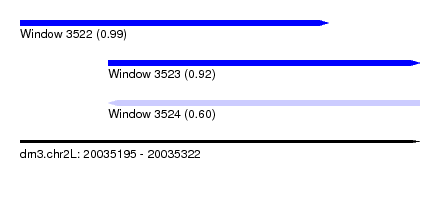
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 20,035,195 – 20,035,322 |
| Length | 127 |
| Max. P | 0.990401 |

| Location | 20,035,195 – 20,035,293 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 69.62 |
| Shannon entropy | 0.58430 |
| G+C content | 0.28381 |
| Mean single sequence MFE | -14.33 |
| Consensus MFE | -9.40 |
| Energy contribution | -10.57 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990401 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
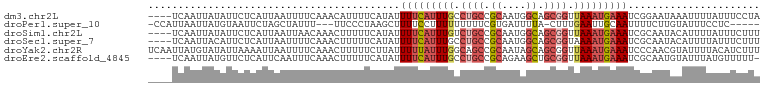
>dm3.chr2L 20035195 98 + 23011544 ----UCAAUUAUAUUCUCAUUAAUUUUCAAACAUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGGAAUAAAUUUUAUUUCCUA ----......................................(((((((((.((((.((....)).)))).)))))))))..((((..........)))).. ( -17.80, z-score = -2.05, R) >droPer1.super_10 756623 92 - 3432795 -CCAUUAAUUAUGUAAUUCUAGCUAUUU---UUCCCUAAGCUUUUCCUUUUUUUUUCGUGAUUUUA-CUUUGAAUUGCAAUUUUCUUGUAUUUCCUC----- -..........((((((((.((((....---.......))))...............(((....))-)...))))))))..................----- ( -8.80, z-score = -2.00, R) >droSim1.chr2L 19690123 98 + 22036055 ----UCAAUUAUAUUCUCAUUAAUUAACAAACUUUUUCAUAUUUUCAUUUGUCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACAUUUUAUUUCUUU ----......................................(((((((((.((((.((....)).)))).)))))))))...................... ( -15.10, z-score = -2.04, R) >droSec1.super_7 3646147 98 + 3727775 ----UCAAUUACAUUCUCAUUAAUUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUAAAAUGAAAUCGCAAUACAUUUUAUUUCUUU ----......................................((((((((..((((.((....)).))))..))))))))...................... ( -14.30, z-score = -1.41, R) >droYak2.chr2R 6498537 102 + 21139217 UCAAUUAUGUAUAUUAAAAUUAAUUUUCAAACUUUUUCUUAUUUUUAUUUGGCAGCCGCAAUAGCAGCGGUUAAAUGAAAUCCCAACGUAUUUUACAUCUUU .....(((((..((((....))))..................(((((((((((.((.((....)).)).))))))))))).....)))))............ ( -13.50, z-score = -0.76, R) >droEre2.scaffold_4845 18292846 97 - 22589142 ----UCAAUUAUGUUCUCAUUCAAUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAGAAGCUGCGGUUAAAUGAAAUCGCAAUGUAUUUAUGUUUUU- ----........................((((.....(((..(((((((((...(((((((...)))))))))))))))).....))).......))))..- ( -16.50, z-score = -1.12, R) >consensus ____UCAAUUAUAUUCUCAUUAAUUUUCAAACUUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUAUAUUUUAUUUCUUU ..........................................(((((((((.((((.((....)).)))).)))))))))...................... ( -9.40 = -10.57 + 1.17)

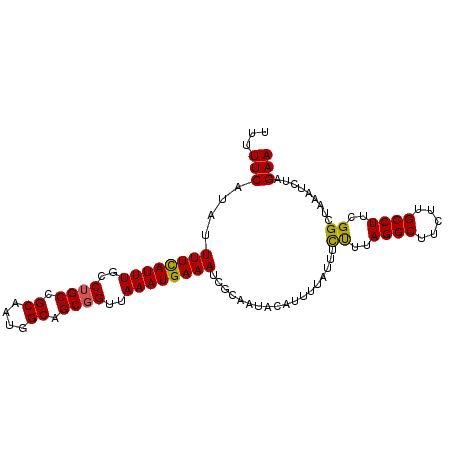
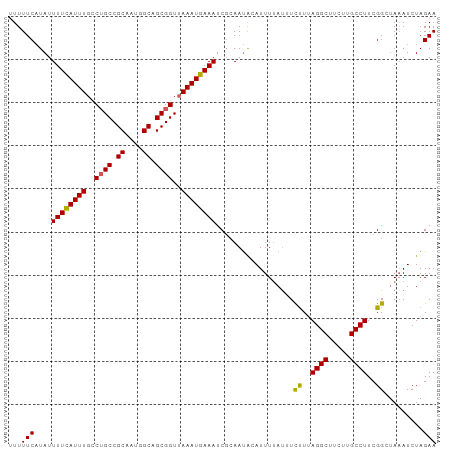
| Location | 20,035,223 – 20,035,322 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.58 |
| Shannon entropy | 0.22489 |
| G+C content | 0.36312 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -20.46 |
| Energy contribution | -20.18 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
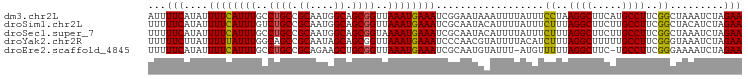
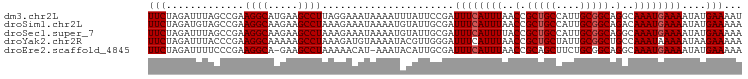
>dm3.chr2L 20035223 99 + 23011544 AUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGGAAUAAAUUUUAUUUCCUAAGGCUUCAUGCCUUCGGCUAAAUCUAGAA ...(((....(((((((((.((((.((....)).)))).)))))))))...)))............((.(((((.....))))).))............ ( -25.60, z-score = -2.10, R) >droSim1.chr2L 19690151 99 + 22036055 UUUUUCAUAUUUUCAUUUGUCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACAUUUUAUUUCUUUAGGCUUCUUGCCUUCGGCUACAUCUAGAA ...(((....(((((((((.((((.((....)).)))).)))))))))..................((..((((.....))))..)).........))) ( -21.30, z-score = -1.77, R) >droSec1.super_7 3646175 99 + 3727775 UUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUAAAAUGAAAUCGCAAUACAUUUUAUUUCUUUAGGCUUCUUGCCUUCGGCUAAAUCUAGAA .........(((..(((((((((((((.......))))))(((((((((........)))))))))....((((.....))))..))).))))..))). ( -21.10, z-score = -1.11, R) >droYak2.chr2R 6498569 99 + 21139217 UUUUUCUUAUUUUUAUUUGGCAGCCGCAAUAGCAGCGGUUAAAUGAAAUCCCAACGUAUUUUACAUCUUUAGGCUUUUUGCCUUCGGGUAAAUCUAGAA ..........(((((((((((.((.((....)).)).)))))))))))...................((((((...((((((....)))))))))))). ( -24.00, z-score = -2.21, R) >droEre2.scaffold_4845 18292874 97 - 22589142 UUUUUCAUAUUUUCAUUUGCCUGCCGCAGAAGCUGCGGUUAAAUGAAAUCGCAAUGUAUUU-AUGUUUUUAGGCUUC-UGCCUUCGGGAAAAUCUAGAA .........(((..((((.((((..(((((((((((((((......))))))(((((....-)))))....))))))-)))...)))).))))..))). ( -25.90, z-score = -1.99, R) >consensus UUUUUCAUAUUUUCAUUUGCCUGCCGCAAUGGCAGCGGUUAAAUGAAAUCGCAAUACAUUUUAUUUCUUUAGGCUUCUUGCCUUCGGCUAAAUCUAGAA ...(((....((((((((..((((.((....)).))))..))))))))..................((..((((.....))))..)).........))) (-20.46 = -20.18 + -0.28)
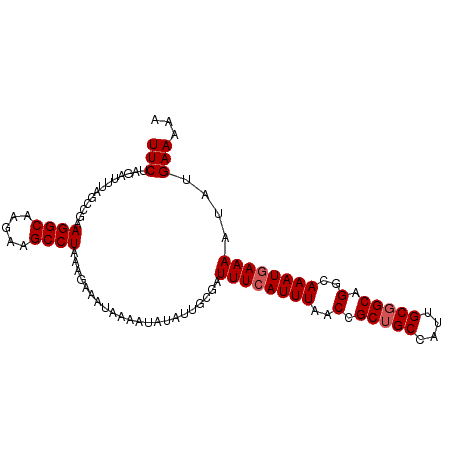
| Location | 20,035,223 – 20,035,322 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Shannon entropy | 0.22489 |
| G+C content | 0.36312 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 20035223 99 - 23011544 UUCUAGAUUUAGCCGAAGGCAUGAAGCCUUAGGAAAUAAAAUUUAUUCCGAUUUCAUUUAACCGCUGCCAUUGCGGCAGGCAAAUGAAAAUAUGAAAAU ((((((((((..((.(((((.....))))).)).....)))))))......((((((((..(.(((((....))))).)..))))))))....)))... ( -25.80, z-score = -2.27, R) >droSim1.chr2L 19690151 99 - 22036055 UUCUAGAUGUAGCCGAAGGCAAGAAGCCUAAAGAAAUAAAAUGUAUUGCGAUUUCAUUUAACCGCUGCCAUUGCGGCAGACAAAUGAAAAUAUGAAAAA (((.............((((.....))))......................((((((((.....(((((.....)))))..))))))))....)))... ( -18.70, z-score = -0.83, R) >droSec1.super_7 3646175 99 - 3727775 UUCUAGAUUUAGCCGAAGGCAAGAAGCCUAAAGAAAUAAAAUGUAUUGCGAUUUCAUUUUACCGCUGCCAUUGCGGCAGGCAAAUGAAAAUAUGAAAAA ...........(((..((((.....)))).......(((((((...........)))))))..(((((....))))).))).................. ( -20.60, z-score = -0.64, R) >droYak2.chr2R 6498569 99 - 21139217 UUCUAGAUUUACCCGAAGGCAAAAAGCCUAAAGAUGUAAAAUACGUUGGGAUUUCAUUUAACCGCUGCUAUUGCGGCUGCCAAAUAAAAAUAAGAAAAA ((((.((..(.(((..((((.....))))...((((((...))))))))))..))........(((((....)))))...............))))... ( -22.90, z-score = -2.10, R) >droEre2.scaffold_4845 18292874 97 + 22589142 UUCUAGAUUUUCCCGAAGGCA-GAAGCCUAAAAACAU-AAAUACAUUGCGAUUUCAUUUAACCGCAGCUUCUGCGGCAGGCAAAUGAAAAUAUGAAAAA (((...((((((....((((.-...))))........-.......(((((....)......((((((...))))))...))))..))))))..)))... ( -21.10, z-score = -1.56, R) >consensus UUCUAGAUUUAGCCGAAGGCAAGAAGCCUAAAGAAAUAAAAUAUAUUGCGAUUUCAUUUAACCGCUGCCAUUGCGGCAGGCAAAUGAAAAUAUGAAAAA (((.............((((.....))))......................((((((((..(.(((((....))))).)..))))))))....)))... (-17.64 = -18.04 + 0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:38 2011