
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,853,535 – 1,853,607 |
| Length | 72 |
| Max. P | 0.620914 |

| Location | 1,853,535 – 1,853,607 |
|---|---|
| Length | 72 |
| Sequences | 15 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.55933 |
| G+C content | 0.54224 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.53 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
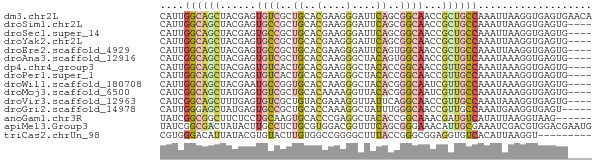
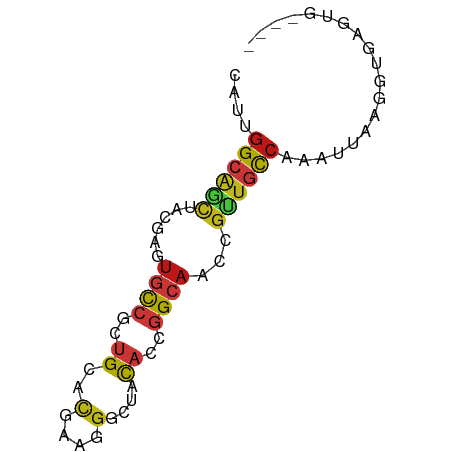
>dm3.chr2L 1853535 72 + 23011544 CAUUGGCAGCUACGAGUGUCGCUGCACGAAGGGAUUCAGCGGCAACCGCUGCCAAAUUAAGGUGAGUGAACA ..((((((((......((((((((..(....)....))))))))...))))))))................. ( -27.10, z-score = -2.37, R) >droSim1.chr2L 1840714 68 + 22036055 CAUUGGCAGCUACGAGUGCCGCUGCACGAAGGGAUUCAGCGGCAACCGCUGCCAAAUUAAGGUGAGUG---- ..((((((((......((((((((..(....)....))))))))...)))))))).............---- ( -29.80, z-score = -3.34, R) >droSec1.super_14 1800238 68 + 2068291 CAUUGGCAGCUACGAGUGCCGCUGCACGAAGGGAUUCAGCGGCAACCGCUGCCAAAUUAAGGUGAGUG---- ..((((((((......((((((((..(....)....))))))))...)))))))).............---- ( -29.80, z-score = -3.34, R) >droYak2.chr2L 1827327 68 + 22324452 CAUUGGCAGCUACGAGUGCCGCUGCACGAAGGGAUUCAGCGGCAACCGCUGCCAAAUUAAGGUGAGUG---- ..((((((((......((((((((..(....)....))))))))...)))))))).............---- ( -29.80, z-score = -3.34, R) >droEre2.scaffold_4929 1900440 68 + 26641161 CAUUGGCAGCUACGAGUGCCGCUGCACGAAGGGAUUCAGUGGCAACCGCUGCCAAAUUAAGGUGAGUG---- ..((((((((......((((((((..(....)....))))))))...)))))))).............---- ( -27.90, z-score = -2.84, R) >droAna3.scaffold_12916 446537 68 - 16180835 CAUCGGCAGCUACGAGUGUCGCUGCACCAAGGGCUACAGUGGCAACCGCUGUCAAAUAAAGGUGAGUG---- ((((.(((((.((....)).)))))..........(((((((...)))))))........))))....---- ( -20.40, z-score = -0.37, R) >dp4.chr4_group3 8586969 68 + 11692001 CAUUGGCAGCUACGAGUGUCACUGCACGAAGGGCUACACCGGCAACCGUUGCCAAAUAAAGGUGAGUG---- ..((((((((...(.((((....((.(....))).)))))(....).)))))))).............---- ( -18.90, z-score = -0.20, R) >droPer1.super_1 10040885 68 + 10282868 CAUUGGCAGCUACGAGUGUCACUGCACGAAGGGCUACACCGGCAACCGUUGCCAAAUAAAGGUGAGUG---- ..((((((((...(.((((....((.(....))).)))))(....).)))))))).............---- ( -18.90, z-score = -0.20, R) >droWil1.scaffold_180708 751369 68 - 12563649 CAUUGGCAGCUACGAAUGCCGGUGCACCAAGGGCUACACGGGCAAUCGUUGCCAAAUAAAGGUGAGUG---- ..((((((...((((.((((.(((..(....)....))).)))).)))))))))).............---- ( -23.40, z-score = -1.67, R) >droMoj3.scaffold_6500 7288148 68 + 32352404 CAUCGGCAGCUAUGAGUGUCGCUGCACAAAAGGUUACACGGGCAAUCGUUGCCAAAUAAAGGUGAGUG---- ((((.(((((.((....)).)))))......(((...((((....)))).))).......))))....---- ( -17.20, z-score = -0.44, R) >droVir3.scaffold_12963 7431304 68 - 20206255 CAUCGGCAGCUUUGAGUGUCGCUGUACGAAAGGUUAUUCAGGCAACCGUUGCCAAAUAAAGGUGAGUG---- ((((.(((((..........))))).(....)........((((.....)))).......))))....---- ( -15.40, z-score = 0.01, R) >droGri2.scaffold_14978 892324 67 + 1124632 CAUUGGGAGCUAUGAGUGCCGCUGCACCAAAGGCUAUUUGGGCAACCGUUGCCAAAUGAAGGUGAGU----- ..((((.(((..((.((((....))))))..........((....))))).))))............----- ( -17.30, z-score = 0.55, R) >anoGam1.chr3R 4295507 66 - 53272125 UAUCGGCGGCUUCUCCUGCAAGUGCACCCGAGGCUACACCGGCAAACGAUGUCAUAUUAAGGUAAG------ ((((((.((((((...(((....)))...))))))...))((((.....)))).......))))..------ ( -16.10, z-score = -0.19, R) >apiMel3.Group3 2813000 72 + 12341916 UAUCGGCGACUAUACUUGCCUCUGCGUGGACGGUUUCAGCGGGAAACAUUGCGAAAUCGACGUGGACGAAUG ..(((((((......)))))((..(((...(((((((.((.(....)...))))))))))))..)).))... ( -23.60, z-score = -2.13, R) >triCas2.chrUn_98 113507 63 - 186310 CGUGGGACAUUAUACGUGUACUUGUGGCCGGGGCUUUACCGGGCGGAGGUGUCACAUUAAGGU--------- .(((........)))((((((((.((.((((.......)))).)).))))).)))........--------- ( -20.80, z-score = -1.52, R) >consensus CAUUGGCAGCUACGAGUGCCGCUGCACGAAGGGCUACACCGGCAACCGUUGCCAAAUUAAGGUGAGUG____ ....((((((......((((..((..(....)....))..))))...))))))................... ( -9.56 = -9.53 + -0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:31 2011