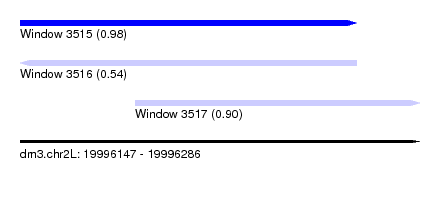
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,996,147 – 19,996,286 |
| Length | 139 |
| Max. P | 0.977442 |

| Location | 19,996,147 – 19,996,264 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.25 |
| Shannon entropy | 0.46678 |
| G+C content | 0.52986 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
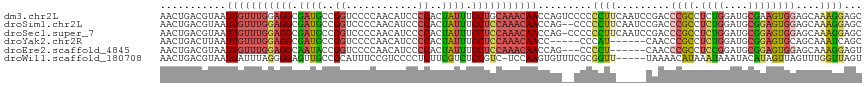
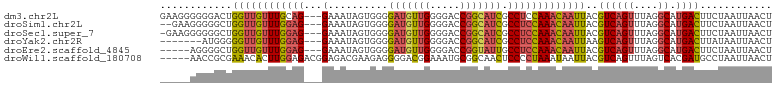
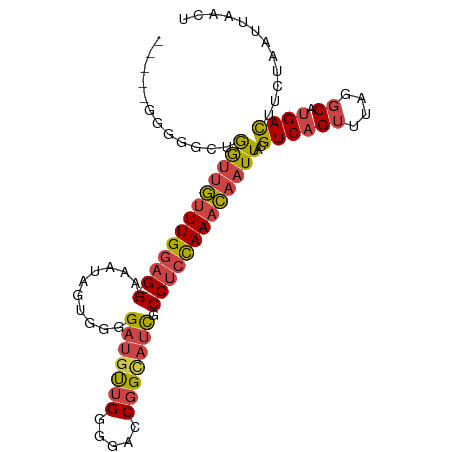
>dm3.chr2L 19996147 117 + 23011544 GCUCCUUUGCUCCACUUCGCAUCCAGAGGCGGGUCGGAUUGAAGGGGGGACUGGUUGUUUGCAGGAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUU .(((((((..((((((.(((........)))))).)))..))))))).(((((((((((((.........(.((.((((((((...)))))))).)).))))))))))).))).... ( -43.10, z-score = -1.02, R) >droSim1.chr2L 19656690 115 + 22036055 GCUCCUUUGCUCCACUCCGCAUCCAGAGGCGGGUCGGAUUGAAGGGGG--CUGGUUGUUUGGAGGAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUU .(((((((..(((((.((((........)))))).)))..)))))))(--(((((((((((((((..........((((((((...)))))))).)))))))))))))).))..... ( -48.40, z-score = -2.34, R) >droSec1.super_7 3607511 116 + 3727775 GCUCCUUUGCUCCACUCCGCAUCCAGAGGCGGGUCGGAUUGAAGGGGGG-CUGGUUGUUUGGAGGAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUU .(((((((..(((((.((((........)))))).)))..)))))))((-(((((((((((((((..........((((((((...)))))))).)))))))))))))).))).... ( -51.60, z-score = -2.96, R) >droYak2.chr2R 6465355 106 + 21139217 GCUGAUUUGCUGCACUCCGCAUCCAGAGGCGGGUUG------AUGGG-----GGUUGUUUGGAGGAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUAAGUCAGUU (((((((((.....((((((((((......))).))------.))))-----)((((((((((((..........((((((((...)))))))).))))))))))))))))))))). ( -42.30, z-score = -2.53, R) >droEre2.scaffold_4845 18257463 108 - 22589142 ACUCCUUUGCUCCACUCCGCAUCCGGAGGCGGGUUG------AGGGG---CUGGUUGUUUGGAGGAAAUAGUGGGGAUGUUGGGGACCGGUAUUGCCUCCAAACAAUUACGUCAGUU ...((((.((((..(((((....)))))..)))).)------)))((---(((((((((((((((.((((.(((............))).)))).)))))))))))))).))).... ( -47.20, z-score = -2.99, R) >droWil1.scaffold_180708 8900069 111 - 12563649 ACUAACCAAACUAACUAUGUAUUUAUUUAUGUUUUA-----AACCGCGAAACACUUGGA-GACGGAGACGAAGAGGGGACGGAAAUGCGGCAACUCCCCUAAAUAAUUACGUCAGUU ............(((((((((.((((((.((((((.-----......))))))((((..-..)(....).)))((((((.(......)(....))))))))))))).))))).)))) ( -23.30, z-score = -0.94, R) >consensus GCUCCUUUGCUCCACUCCGCAUCCAGAGGCGGGUCG_____AAGGGGG__CUGGUUGUUUGGAGGAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUU .(((((.........(((((........))))).........))))).....(((((((((((((..........(((((((.....))))))).)))))))))))))......... (-24.80 = -25.67 + 0.87)


| Location | 19,996,147 – 19,996,264 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Shannon entropy | 0.46678 |
| G+C content | 0.52986 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19996147 117 - 23011544 AACUGACGUAAUUGUUUGGAGGCGAUGCCGGUCCCCAACAUCCCCACUAUUUCCUGCAAACAACCAGUCCCCCCUUCAAUCCGACCCGCCUCUGGAUGCGAAGUGGAGCAAAGGAGC ....(((....(((((((.(((.((((..((............))..)))).))).)))))))...)))...((((...((((...(((........)))...))))...))))... ( -28.30, z-score = 0.22, R) >droSim1.chr2L 19656690 115 - 22036055 AACUGACGUAAUUGUUUGGAGGCGAUGCCGGUCCCCAACAUCCCCACUAUUUCCUCCAAACAACCAG--CCCCCUUCAAUCCGACCCGCCUCUGGAUGCGGAGUGGAGCAAAGGAGC ...........(((((((((((.((((..((............))..)))).)))))))))))...(--(..((((...((((..((((........))))..))))...)))).)) ( -35.00, z-score = -1.57, R) >droSec1.super_7 3607511 116 - 3727775 AACUGACGUAAUUGUUUGGAGGCGAUGCCGGUCCCCAACAUCCCCACUAUUUCCUCCAAACAACCAG-CCCCCCUUCAAUCCGACCCGCCUCUGGAUGCGGAGUGGAGCAAAGGAGC ...........(((((((((((.((((..((............))..)))).)))))))))))....-....((((...((((..((((........))))..))))...))))... ( -34.50, z-score = -1.30, R) >droYak2.chr2R 6465355 106 - 21139217 AACUGACUUAAUUGUUUGGAGGCGAUGCCGGUCCCCAACAUCCCCACUAUUUCCUCCAAACAACC-----CCCAU------CAACCCGCCUCUGGAUGCGGAGUGCAGCAAAUCAGC ..((((.....(((((((((((.((((..((............))..)))).)))))))))))..-----.....------......(((((((....))))).))......)))). ( -30.20, z-score = -2.42, R) >droEre2.scaffold_4845 18257463 108 + 22589142 AACUGACGUAAUUGUUUGGAGGCAAUACCGGUCCCCAACAUCCCCACUAUUUCCUCCAAACAACCAG---CCCCU------CAACCCGCCUCCGGAUGCGGAGUGGAGCAAAGGAGU ...........(((((((((((.((((..((............))..)))).)))))))))))...(---(.(((------...(((((.((((....)))))))).)...))).)) ( -31.80, z-score = -1.65, R) >droWil1.scaffold_180708 8900069 111 + 12563649 AACUGACGUAAUUAUUUAGGGGAGUUGCCGCAUUUCCGUCCCCUCUUCGUCUCCGUC-UCCAAGUGUUUCGCGGUU-----UAAAACAUAAAUAAAUACAUAGUUAGUUUGGUUAGU ((((((((((.((((((((((((...............))))).........((((.-............))))..-----.......))))))).)))...)))))))........ ( -19.48, z-score = 0.24, R) >consensus AACUGACGUAAUUGUUUGGAGGCGAUGCCGGUCCCCAACAUCCCCACUAUUUCCUCCAAACAACCAG__CCCCCUU_____CAACCCGCCUCUGGAUGCGGAGUGGAGCAAAGGAGC ...........(((((((((((.((((..((............))..)))).))))))))))).........((((.....((..((((........))))..)).....))))... (-17.92 = -18.12 + 0.20)
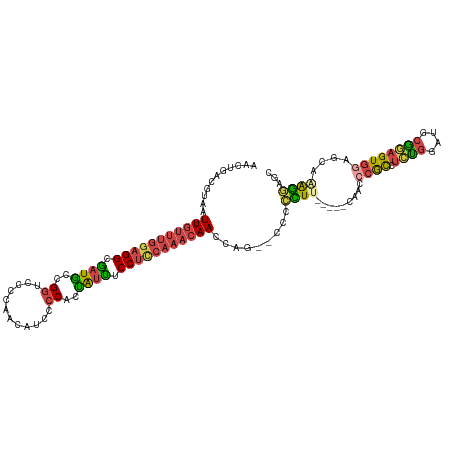
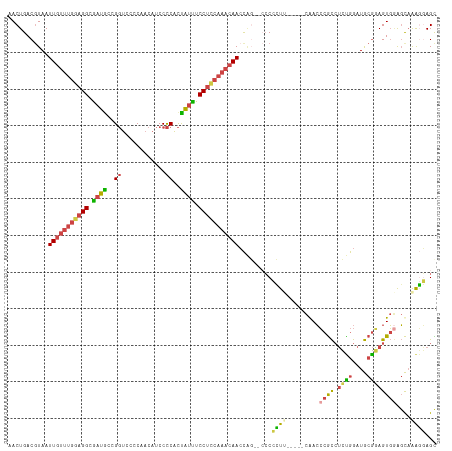
| Location | 19,996,187 – 19,996,286 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Shannon entropy | 0.38934 |
| G+C content | 0.47275 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -18.57 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19996187 99 + 23011544 GAAGGGGGGACUGGUUGUUUGCAG---GAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUUUAGGCAUGACUUCUAAUUAACU ...((((((((((((((((((...---......(.((.((((((((...)))))))).)).))))))))))).))).((....))....)))))........ ( -28.00, z-score = -0.78, R) >droSim1.chr2L 19656730 97 + 22036055 --GAAGGGGGCUGGUUGUUUGGAG---GAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUUUAGGCAUGACUUCUAAUUAACU --((((..((((((((((((((((---(..........((((((((...)))))))).)))))))))))))).))).((....))....))))......... ( -33.40, z-score = -2.72, R) >droSec1.super_7 3607551 98 + 3727775 -GAAGGGGGGCUGGUUGUUUGGAG---GAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUUUAGGCAUGACUUCUAAUUAACU -..(((((((((((((((((((((---(..........((((((((...)))))))).)))))))))))))).))).((....))....)))))........ ( -33.50, z-score = -2.61, R) >droYak2.chr2R 6465391 92 + 21139217 -------AUGGGGGUUGUUUGGAG---GAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUAAGUCAGUUUAGGCAUGACUUAUAAUUAACU -------......(((((((((((---(..........((((((((...)))))))).))))))))))))(((((((((....)).)))))))......... ( -31.80, z-score = -3.54, R) >droEre2.scaffold_4845 18257499 94 - 22589142 -----AGGGGCUGGUUGUUUGGAG---GAAAUAGUGGGGAUGUUGGGGACCGGUAUUGCCUCCAAACAAUUACGUCAGUUUAGGCAUGACUUCUAAUUAACU -----(((((.(((((((((((((---(.((((.(((............))).)))).)))))))))))))).(((......)))....)))))........ ( -31.00, z-score = -2.51, R) >droWil1.scaffold_180708 8900105 97 - 12563649 -----AACCGCGAAACACUUGGAGACGGAGACGAAGAGGGGACGGAAAUGCGGCAACUCCCCUAAAUAAUUACGUCAGUUUAGUCACGAUGCCUAAUUAACU -----...((.(((((..(((....))).((((((.((((((.(......)(....)))))))......)).)))).)))...)).)).............. ( -18.10, z-score = 0.79, R) >consensus _____GGGGGCUGGUUGUUUGGAG___GAAAUAGUGGGGAUGUUGGGGACCGGCAUCGCCUCCAAACAAUUACGUCAGUUUAGGCAUGACUUCUAAUUAACU ............(((((((((((............((.(((((((.....))))))).)))))))))))))..((((((....)).))))............ (-18.57 = -19.35 + 0.78)
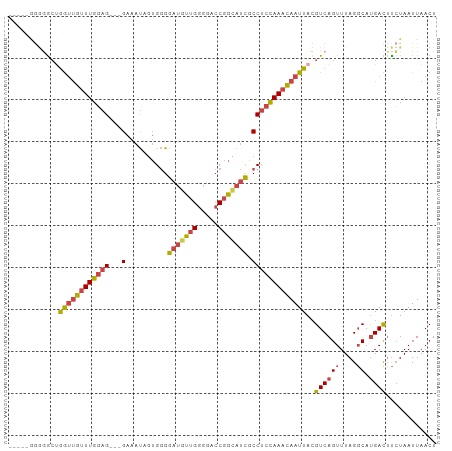
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:33 2011