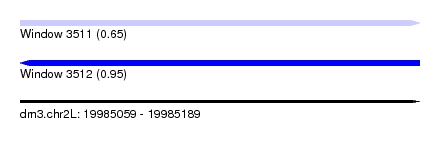
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,985,059 – 19,985,189 |
| Length | 130 |
| Max. P | 0.953955 |

| Location | 19,985,059 – 19,985,189 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 130 |
| Reading direction | forward |
| Mean pairwise identity | 86.70 |
| Shannon entropy | 0.23874 |
| G+C content | 0.46744 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -27.46 |
| Energy contribution | -29.10 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
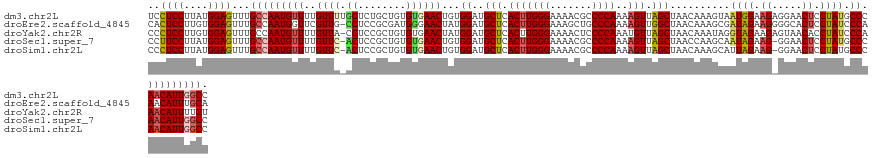
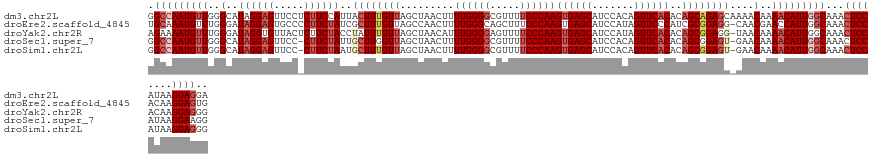
>dm3.chr2L 19985059 130 + 23011544 UCCUCCUUAUGGAGUUUGCCAAUGUUUUGUUUUGCUCUGCUGUGUGAACUGUGGAUGCUCACUUGGGAAAACGCCCCAAAAGUUAGCUAACAAAGUAAUGGAAGAGGAACUCCUAUGCCCAACAUUGGCC ..((((....))))...((((((((((((((..(((..(((..((((.(.......).))))(((((.......))))).))).))).))))))....(((...(((....)))....))))))))))). ( -39.10, z-score = -2.14, R) >droEre2.scaffold_4845 18246852 129 - 22589142 CACUCCUUGUGGAGUUUGCCAAUGGUUCGUUG-CCUCCGCGAUGGGAACUAUGGAUGCUCACUUGGGAAAGCUGCCCAAAAGUUGGCUAACAAAGCGAUAGAAGGGGCACUCCUAUCCCAAACAUUUGCA (((.....)))((((...(((.((((((((((-(....)))))..)))))))))..))))..((((((.((.(((((......(.(((.....))).)......)))))...)).))))))......... ( -41.60, z-score = -0.95, R) >droYak2.chr2R 6454566 129 + 21139217 CCCUCCUUGUGGAGUUUGCCAAUGUUUUGUUA-CCUCCGCUGUGUGAACUAUGGAUGCUCACUUGGGAAAACUCCCCAAAUGUUAGCUAACAAAUAGGUAGAAGAGUAACACCUAUCCCAAACAUUUUCU ..((((....))))......((((((((((((-..((((..((....))..)))).(((.(((((((.......)))))..)).)))))))).((((((.(........)))))))...))))))).... ( -28.10, z-score = -0.46, R) >droSec1.super_7 3596512 128 + 3727775 CCUUCCUUAUGGAGUUUGCCAAUGUUUUGUUC-ACUCCGCUGUGUGAACUGUGGAUGCUCACUUGGGAAAACGCCCCAAAAGUUAGCUAACCAAGCAAUAGAAG-GGAACUCCUAUGCCCAACAUUGGCC ..((((....))))...(((((((((..((((-((........))))))..(((..(((.(((((((.......))))..))).)))...))).(((.(((.((-....)).))))))..))))))))). ( -41.40, z-score = -2.80, R) >droSim1.chr2L 19645679 128 + 22036055 CCCUCCUUAUGGAGUUUGCCAAUGUUUUGUUC-ACUCCGCUGUGUGAACUGUGGAUGCUCACUUGGGAAAACGCCCCAAAAGUUAGCUAACAAAGCAUUAGAAG-GGAACUCCUAUGCCCAACAUUGGCC ..((((....))))...(((((((((..((((-((........))))))...((..(((.(((((((.......))))..))).))).......((((.((.((-....)).))))))))))))))))). ( -42.70, z-score = -3.18, R) >consensus CCCUCCUUAUGGAGUUUGCCAAUGUUUUGUUC_ACUCCGCUGUGUGAACUGUGGAUGCUCACUUGGGAAAACGCCCCAAAAGUUAGCUAACAAAGCAAUAGAAG_GGAACUCCUAUGCCCAACAUUGGCC ..((((....))))...((((((((((((((....(((((.((....)).))))).(((.(((((((.......))))..))).))).))))))...((((.((.....)).)))).....)))))))). (-27.46 = -29.10 + 1.64)
| Location | 19,985,059 – 19,985,189 |
|---|---|
| Length | 130 |
| Sequences | 5 |
| Columns | 130 |
| Reading direction | reverse |
| Mean pairwise identity | 86.70 |
| Shannon entropy | 0.23874 |
| G+C content | 0.46744 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -31.54 |
| Energy contribution | -33.82 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
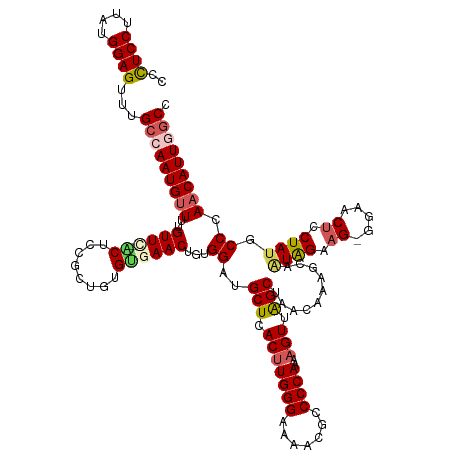
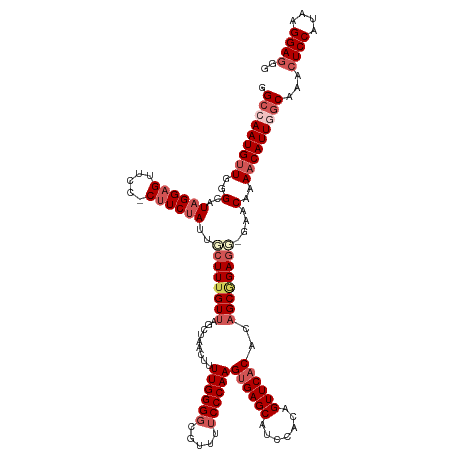
>dm3.chr2L 19985059 130 - 23011544 GGCCAAUGUUGGGCAUAGGAGUUCCUCUUCCAUUACUUUGUUAGCUAACUUUUGGGGCGUUUUCCCAAGUGAGCAUCCACAGUUCACACAGCAGAGCAAAACAAAACAUUGGCAAACUCCAUAAGGAGGA .((((((((((((...(((....)))..))).....((((((.(((.....((((((.....))))))((((((.......))))))..)))..))))))....)))))))))...((((....)))).. ( -43.70, z-score = -3.23, R) >droEre2.scaffold_4845 18246852 129 + 22589142 UGCAAAUGUUUGGGAUAGGAGUGCCCCUUCUAUCGCUUUGUUAGCCAACUUUUGGGCAGCUUUCCCAAGUGAGCAUCCAUAGUUCCCAUCGCGGAGG-CAACGAACCAUUGGCAAACUCCACAAGGAGUG (((.((((...(.((((((((.....)))))))).)((((((.(((.(((..((((..((((........)))).)))).)))(((......)))))-))))))).)))).))).(((((....))))). ( -39.90, z-score = -0.97, R) >droYak2.chr2R 6454566 129 - 21139217 AGAAAAUGUUUGGGAUAGGUGUUACUCUUCUACCUAUUUGUUAGCUAACAUUUGGGGAGUUUUCCCAAGUGAGCAUCCAUAGUUCACACAGCGGAGG-UAACAAAACAUUGGCAAACUCCACAAGGAGGG .(..(((((((.(((((((((.........)))))))))((((.((.....(((((((...)))))))((((((.......)))))).......)).-)))).)))))))..)...((((....)))).. ( -38.20, z-score = -2.04, R) >droSec1.super_7 3596512 128 - 3727775 GGCCAAUGUUGGGCAUAGGAGUUCC-CUUCUAUUGCUUGGUUAGCUAACUUUUGGGGCGUUUUCCCAAGUGAGCAUCCACAGUUCACACAGCGGAGU-GAACAAAACAUUGGCAAACUCCAUAAGGAAGG .((((((((((((((((((((....-)))))).)))))((...(((....(((((((.....)))))))..)))..))...((((((........))-))))..)))))))))....(((....)))... ( -44.90, z-score = -2.76, R) >droSim1.chr2L 19645679 128 - 22036055 GGCCAAUGUUGGGCAUAGGAGUUCC-CUUCUAAUGCUUUGUUAGCUAACUUUUGGGGCGUUUUCCCAAGUGAGCAUCCACAGUUCACACAGCGGAGU-GAACAAAACAUUGGCAAACUCCAUAAGGAGGG .((((((((((((((((((((....-))))).)))).......(((....(((((((.....)))))))..)))..))...((((((........))-))))..)))))))))...((((....)))).. ( -47.30, z-score = -3.44, R) >consensus GGCCAAUGUUGGGCAUAGGAGUUCC_CUUCUAUUGCUUUGUUAGCUAACUUUUGGGGCGUUUUCCCAAGUGAGCAUCCACAGUUCACACAGCGGAGG_GAACAAAACAUUGGCAAACUCCAUAAGGAGGG .(((((((((..(..((((((.....))))))...(((((((.........((((((.....))))))((((((.......))))))..))))))).....)..)))))))))...((((....)))).. (-31.54 = -33.82 + 2.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:29 2011