| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,957,684 – 19,957,776 |
| Length | 92 |
| Max. P | 0.949176 |

| Location | 19,957,684 – 19,957,776 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 66.59 |
| Shannon entropy | 0.68402 |
| G+C content | 0.35761 |
| Mean single sequence MFE | -11.66 |
| Consensus MFE | -10.18 |
| Energy contribution | -10.04 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.44 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949176 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
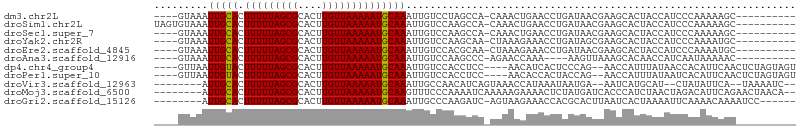
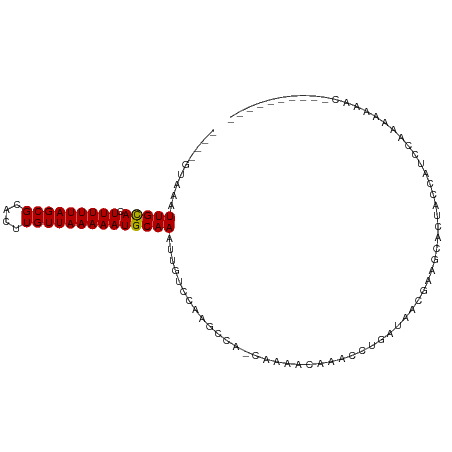
>dm3.chr2L 19957684 92 + 23011544 ----GUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCUAGCCA-CAAACUGAACCUGAUAACGAAGCACUACCAUCCCAAAAAGC---------- ----((...(((((.(((((((((....)))))))))))))).(((((.....((-.....)).....)))))....))..................---------- ( -10.70, z-score = 0.28, R) >droSim1.chr2L 19618436 96 + 22036055 UAGUGUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCAAGCCA-CAAACUGAACCUGAUAACGAAGCACUACCAUCCCAAAAAGC---------- ((((((...(((((.(((((((((....)))))))))))))).(((((.....((-.....)).....)))))....))))))..............---------- ( -16.70, z-score = -1.29, R) >droSec1.super_7 3569211 92 + 3727775 ----GUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCAAGCCA-CAAACUGAACCUGAUAACGAAGCACUACCAUCCCAAAAAGC---------- ----((...(((((.(((((((((....)))))))))))))).(((((.....((-.....)).....)))))....))..................---------- ( -10.70, z-score = 0.34, R) >droYak2.chr2R 6427235 92 + 21139217 ----GUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCAAGCAA-CUAAAGAAACCUGAUAGCGAAGCACUACCAUCCCAAAAUGC---------- ----(((..(((((.(((((((((....)))))))))))))).........((..-(((.((....))..)))....))..))).............---------- ( -11.70, z-score = 0.52, R) >droEre2.scaffold_4845 18219828 92 - 22589142 ----GUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCACGCAA-CUAAAGAAACCUGAUAACGAAGCACUACCAUCCCAAAAUGC---------- ----((...(((((.(((((((((....)))))))))))))).((((....))))-.....................))..................---------- ( -10.80, z-score = 0.25, R) >droAna3.scaffold_12916 7322228 88 - 16180835 ----GUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCAAGCCC-AGAACCAAA----AAGUUAAAGCACAACCAUCAAUAAAAAC---------- ----.....(((((.(((((((((....)))))))))))))).........((..-..(((....----..)))...))..................---------- ( -12.10, z-score = -1.00, R) >dp4.chr4_group4 2805499 97 - 6586962 ----GUUAAUUGUACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCACCUCC----AACAUCACUCCCAG--AACCAUUUAUAACCACAUUCAACUCUAGUAGU ----.....(((((.(((((((((....)))))))))))))).............----..............--................................ ( -8.40, z-score = -0.23, R) >droPer1.super_10 614218 97 - 3432795 ----GUUAAUUGUACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCACCUCC----AACACCACUACCAG--AACCAUUUAUAAUCACAUUCAACUCUAGUAGU ----.....(((((.(((((((((....)))))))))))))).............----......(((((.((--(......................))).))))) ( -12.55, z-score = -1.64, R) >droVir3.scaffold_12963 18946167 91 - 20206255 --------AUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGCCAACAUCAGUAAACCAUAAAUAAUGA--AAUCAUGCAU--CUAUAUUCA--UAAAAUC-- --------.(((((.(((((((((....)))))))))))))).((((........))))..........((((--(.........--.....))))--)......-- ( -11.24, z-score = 0.02, R) >droMoj3.scaffold_6500 4291959 97 - 32352404 --------AUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAGUUUCCCAAAAUCAAAAAGAAAACUCUAUGAUCACCCAUCUAACUAGACAUUCAGAACUAACA-- --------.(((((.(((((((((....))))))))))))))..................(((...((((.(((.....)))....))))..)))..........-- ( -12.40, z-score = -1.34, R) >droGri2.scaffold_15126 7224393 92 + 8399593 --------AUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGCCCAAGAUC-AGUAAGAAACCACGCACUUAAUCACUAAAAUUCAAAACAAAAUCC------ --------.(((((.(((((((((....))))))))))))))..(((......((-.....))......))).............................------ ( -11.00, z-score = -0.75, R) >consensus ____GUAAAUUGCACUUUUUAGCGCACUUGUUAAAAAUGCAAAUUGUCCAAGCCA_CAAAACAAACCUGAUAACGAAGCACUACCAUCCAAAAAAAC__________ .........(((((.(((((((((....))))))))))))))................................................................. (-10.18 = -10.04 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:25 2011