| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,957,542 – 19,957,632 |
| Length | 90 |
| Max. P | 0.991617 |

| Location | 19,957,542 – 19,957,632 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.60454 |
| G+C content | 0.43571 |
| Mean single sequence MFE | -15.48 |
| Consensus MFE | -8.71 |
| Energy contribution | -9.35 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
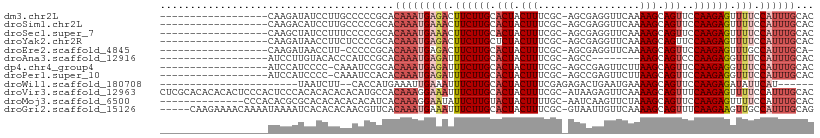
>dm3.chr2L 19957542 90 + 23011544 ------------------CAAGAUAUCCUUGCCCCCGCACAAAUGAGACUUCUUGCACUACUUUCGC-AGCGAGGUUCAAAAGCAGUUCCAAGAGUUUUCCAUUUGCAC ------------------((((.....))))........(((((((((.((((((.(((.((((.(.-(((...)))).)))).)))..)))))).))).))))))... ( -15.60, z-score = -0.04, R) >droSim1.chr2L 19618220 90 + 22036055 ------------------CAAGACAUCCUUGCCCCCGCACAAAUGAAACUUCUUGCACUACUUUCGC-AGCGAGGUUCAAAAGCAGUUCCAAGAGUUUUCCAUUUGCAC ------------------((((.....))))........(((((((((.((((((.(((.((((.(.-(((...)))).)))).)))..)))))).))).))))))... ( -15.50, z-score = -0.21, R) >droSec1.super_7 3569069 90 + 3727775 ------------------CAAGCUAUCCUUUCCCCCGCACAAAUGAAACUUCUUGCACUACUUUCGC-AGCGAGGUUCAAAAGCAGUUCCAAGAGUUUUCCAUUUGCAC ------------------.....................(((((((((.((((((.(((.((((.(.-(((...)))).)))).)))..)))))).))).))))))... ( -14.10, z-score = -0.16, R) >droYak2.chr2R 6427093 90 + 21139217 ------------------CAAGAUAACCUUCUCCCCGCACAAAUGAGACUUCUUGCUCUACUUUCGC-AGCGAGGUUCAAAAGCAGUUCCAAGAGUUUUCCAUUUGCAC ------------------..(((......))).......(((((((((.((((((..((.((((.(.-(((...)))).)))).))...)))))).))).))))))... ( -13.20, z-score = 0.73, R) >droEre2.scaffold_4845 18219688 88 - 22589142 ------------------CAAGAUAACCUU-CCCCCGCACAAAUGAGACUUCUUGCACUACUUUCGC-AGCGAGGUUCAAAAGCAGUUCCAAGAGUUUGCCAUUUGCA- ------------------............-........((((((.((((((((((.........))-)).)))))).....((((..........))))))))))..- ( -14.50, z-score = 0.39, R) >droAna3.scaffold_12916 7322097 82 - 16180835 ------------------AUCCUUGUACACCCAUCCGCACAAAUGAGAUUUCUUGCACUACUUUCGC-AGCC--------AAGCAGUCCCAAGAGGUUUCCAUUUGCAC ------------------.....................((((((((((((((((.(((.(((....-....--------))).)))..)))))))))).))))))... ( -15.90, z-score = -1.83, R) >dp4.chr4_group4 2805357 89 - 6586962 ------------------AUCCAUCCCC-CAAAUCCGCACAAAUGAGAUUUCUUGCACUACUUUCGC-AGCCGAGUUCUUAAGCAGUUCCAAGAGGUUUCCAUUUGCAC ------------------..........-..........((((((((((((((((.(((.(((..((-......))....))).)))..)))))))))).))))))... ( -16.90, z-score = -1.97, R) >droPer1.super_10 614076 89 - 3432795 ------------------AUCCAUCCCC-CAAAUCCACACAAAUGAGAUUUCUUGCACUACUUUCGC-AGCCGAGUUCUUAAGCAGUUCCAAGAGGUUUCCAUUUGCAC ------------------..........-..........((((((((((((((((.(((.(((..((-......))....))).)))..)))))))))).))))))... ( -16.90, z-score = -2.42, R) >droWil1.scaffold_180708 8838692 78 - 12563649 -----------------------UAAUCUU--CACCAUGAAAUUGAAAUUUCUUGCACUACUUUCGAGAGACUGAAUGAAAAGCAGUUCCAAGAGAUAUUCAU------ -----------------------.......--...........((((((((((((.(((.(((((....)).........))).)))..)))))))).)))).------ ( -12.00, z-score = -0.32, R) >droVir3.scaffold_12963 18945771 108 - 20206255 CUCGCACACACACUCCCACUCCCACACACACACAUGCCACAAAGGAAAUUUCUUGCACUACUUUCGC-AUAAGAGUUCAAAAGCAGUUUCAAGAGUUUUCCAUUUGCAC .......................................(((((((((.((((((.(((.((((.((-......))...)))).)))..)))))).))))).))))... ( -15.60, z-score = -1.49, R) >droMoj3.scaffold_6500 4291653 94 - 32352404 --------------CCCACACGCGCACACACACACAUCACAAAGGAAUAUUCUUGUACUACUUUUGC-AAUCAAGUUCUAAAGCAGUUCCAAGAGUUUUCCAUUUGCAC --------------.........................((((((((.(((((((.(((.((((.((-......))...)))).)))..))))))).)))).))))... ( -18.20, z-score = -3.12, R) >droGri2.scaffold_15126 7224215 103 + 8399593 -----CAAGAAAACAAAAUAAAAUCACACACAACGUUCACAAAUGAAAUUUCUUGCACUACUUUCGC-GUAAUUGUUCAAAAGCAGUUUCAAGAAGUUGCCAUUUGCAG -----..................................((((((.(((((((((.(((.((((.((-(....)))...)))).)))..)))))))))..))))))... ( -17.30, z-score = -1.20, R) >consensus __________________CAAGAUAACCUUCACCCCGCACAAAUGAAAUUUCUUGCACUACUUUCGC_AGCCAGGUUCAAAAGCAGUUCCAAGAGUUUUCCAUUUGCAC .......................................(((((((((.((((((.(((.(((.................))).)))..)))))).))).))))))... ( -8.71 = -9.35 + 0.63)

| Location | 19,957,542 – 19,957,632 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 70.64 |
| Shannon entropy | 0.60454 |
| G+C content | 0.43571 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -14.60 |
| Energy contribution | -15.19 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
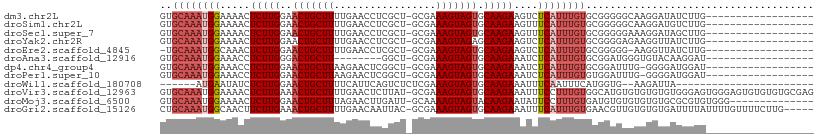
>dm3.chr2L 19957542 90 - 23011544 GUGCAAAUGGAAAACUCUUGGAACUGCUUUUGAACCUCGCU-GCGAAAGUAGUGCAAGAAGUCUCAUUUGUGCGGGGGCAAGGAUAUCUUG------------------ (..((((((((....(((((..((((((((((.........-.)))))))))).)))))..)).))))))..).....((((.....))))------------------ ( -28.30, z-score = -2.12, R) >droSim1.chr2L 19618220 90 - 22036055 GUGCAAAUGGAAAACUCUUGGAACUGCUUUUGAACCUCGCU-GCGAAAGUAGUGCAAGAAGUUUCAUUUGUGCGGGGGCAAGGAUGUCUUG------------------ (..(((((((((...(((((..((((((((((.........-.)))))))))).)))))..)))))))))..).((((((....)))))).------------------ ( -33.90, z-score = -3.82, R) >droSec1.super_7 3569069 90 - 3727775 GUGCAAAUGGAAAACUCUUGGAACUGCUUUUGAACCUCGCU-GCGAAAGUAGUGCAAGAAGUUUCAUUUGUGCGGGGGAAAGGAUAGCUUG------------------ (..(((((((((...(((((..((((((((((.........-.)))))))))).)))))..)))))))))..)..................------------------ ( -29.90, z-score = -2.83, R) >droYak2.chr2R 6427093 90 - 21139217 GUGCAAAUGGAAAACUCUUGGAACUGCUUUUGAACCUCGCU-GCGAAAGUAGAGCAAGAAGUCUCAUUUGUGCGGGGAGAAGGUUAUCUUG------------------ (..((((((((....(((((...(((((((((.........-.)))))))))..)))))..)).))))))..)...((((......)))).------------------ ( -25.80, z-score = -1.32, R) >droEre2.scaffold_4845 18219688 88 + 22589142 -UGCAAAUGGCAAACUCUUGGAACUGCUUUUGAACCUCGCU-GCGAAAGUAGUGCAAGAAGUCUCAUUUGUGCGGGGG-AAGGUUAUCUUG------------------ -.((((((((...(((((((..((((((((((.........-.)))))))))).)))).))).)))))))).(((((.-.......)))))------------------ ( -24.00, z-score = -0.70, R) >droAna3.scaffold_12916 7322097 82 + 16180835 GUGCAAAUGGAAACCUCUUGGGACUGCUU--------GGCU-GCGAAAGUAGUGCAAGAAAUCUCAUUUGUGCGGAUGGGUGUACAAGGAU------------------ (..(((((((...((....))((...(((--------((((-((....))))).))))...)))))))))..)..................------------------ ( -25.00, z-score = -2.04, R) >dp4.chr4_group4 2805357 89 + 6586962 GUGCAAAUGGAAACCUCUUGGAACUGCUUAAGAACUCGGCU-GCGAAAGUAGUGCAAGAAAUCUCAUUUGUGCGGAUUUG-GGGGAUGGAU------------------ (..((((((((....(((((...((.....))......(((-((....))))).)))))..)).))))))..).......-..........------------------ ( -23.40, z-score = -1.16, R) >droPer1.super_10 614076 89 + 3432795 GUGCAAAUGGAAACCUCUUGGAACUGCUUAAGAACUCGGCU-GCGAAAGUAGUGCAAGAAAUCUCAUUUGUGUGGAUUUG-GGGGAUGGAU------------------ ........(....)((((..((.(..(...........(((-((....)))))(((((........))))))..).))..-))))......------------------ ( -20.80, z-score = -0.50, R) >droWil1.scaffold_180708 8838692 78 + 12563649 ------AUGAAUAUCUCUUGGAACUGCUUUUCAUUCAGUCUCUCGAAAGUAGUGCAAGAAAUUUCAAUUUCAUGGUG--AAGAUUA----------------------- ------.((((....(((((..(((((((((.............))))))))).)))))...)))).((((.....)--)))....----------------------- ( -15.52, z-score = -0.56, R) >droVir3.scaffold_12963 18945771 108 + 20206255 GUGCAAAUGGAAAACUCUUGAAACUGCUUUUGAACUCUUAU-GCGAAAGUAGUGCAAGAAAUUUCCUUUGUGGCAUGUGUGUGUGUGGGAGUGGGAGUGUGUGUGCGAG (..((..........(((((..((((((((((.........-.)))))))))).))))).((((((.((.(.((((......)))).).)).))))))...))..)... ( -29.40, z-score = -1.97, R) >droMoj3.scaffold_6500 4291653 94 + 32352404 GUGCAAAUGGAAAACUCUUGGAACUGCUUUAGAACUUGAUU-GCAAAAGUAGUACAAGAAUAUUCCUUUGUGAUGUGUGUGUGUGCGCGUGUGGG-------------- ..(((((.((((.(.(((((..((((((((.(.........-.).)))))))).))))).).))))))))).(..(((((....)))))..)...-------------- ( -25.00, z-score = -2.05, R) >droGri2.scaffold_15126 7224215 103 - 8399593 CUGCAAAUGGCAACUUCUUGAAACUGCUUUUGAACAAUUAC-GCGAAAGUAGUGCAAGAAAUUUCAUUUGUGAACGUUGUGUGUGAUUUUAUUUUGUUUUCUUG----- ..((((((((.((.((((((..((((((((((.........-.)))))))))).)))))).))))))))))(((((..(((........)))..))))).....----- ( -26.00, z-score = -2.09, R) >consensus GUGCAAAUGGAAAACUCUUGGAACUGCUUUUGAACCUCGCU_GCGAAAGUAGUGCAAGAAAUCUCAUUUGUGCGGGGGGAAGGUUAUCUUG__________________ ..((((((((.....(((((..(((((((.................))))))).)))))....))))))))...................................... (-14.60 = -15.19 + 0.59)
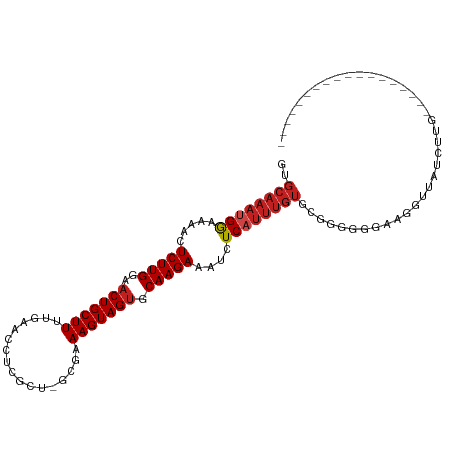
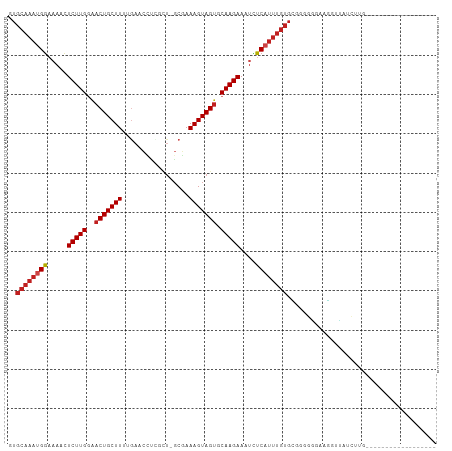
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:24 2011