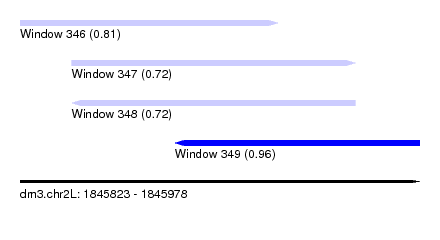
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,845,823 – 1,845,978 |
| Length | 155 |
| Max. P | 0.960271 |

| Location | 1,845,823 – 1,845,923 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.51 |
| Shannon entropy | 0.68646 |
| G+C content | 0.28313 |
| Mean single sequence MFE | -18.31 |
| Consensus MFE | -6.40 |
| Energy contribution | -5.68 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1845823 100 + 23011544 --------------------CUUUUGAAAAAAGCAGUUGUAGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAAA --------------------..........((((........((((........((((((.....))))))...))))..((((((....))))))..........)))).......... ( -16.80, z-score = -1.09, R) >droEre2.scaffold_4929 1892810 119 + 26641161 AUGUUUCUUUUGAAGUUUAAUUUUUAUAAUAAGCAAUAACGGGAGAUGCAAAAUGUAUAAAUAAUUUGCGGUCCUUUAAUAGCCACAGGAGUGGUUAUUUAAUUAAGUUUAGAAAAAAA- ...(((.(((((((.(((((((..................((((..((((((.(((....))).)))))).))))..(((((((((....))))))))).)))))))))))))).))).- ( -24.50, z-score = -2.03, R) >droSec1.super_14 1792558 99 + 2068291 --------------------UUUCUUAAAAAAUCAGUUGUAGCAGAUUUAAAAUCCAUUAAUAAUUUGUGGCCCUCUGAAAACCACAAAUGUGGUUAUUAAUUUAAGCUUAGAAAAAAA- --------------------...((((((...((((..((.(((((((..............))))))).))...)))).((((((....)))))).....))))))............- ( -18.84, z-score = -2.32, R) >droSim1.chr2L 1832914 99 + 22036055 --------------------CUUCUUAAAAAAUCAGUUGUAGCUGAUUUAAAAUCCAUUAAUAAUUUGUGGCCUUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAA- --------------------...((((((((((((((....)))))))).....(((((.....(((((((...........)))))))))))).......))))))............- ( -19.20, z-score = -2.75, R) >dp4.chr4_group3 8578231 100 + 11692001 ----------AUGAUUUCCUCCUUUUACCCAAGUGAUUAUGGGAUUCGUAAUAGUUUCAAAGAAGUCAGUCCUUCCUAUAGAUCGU---UUCUAUCAAUCAUUUCAACCCCAA------- ----------.(((((((..((..((((....))))....))((..(......)..))...))))))).........(((((....---.)))))..................------- ( -12.20, z-score = 0.27, R) >consensus ____________________CUUUUUAAAAAAGCAGUUGUAGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAA_ ................................................................................((((((....))))))........................ ( -6.40 = -5.68 + -0.72)

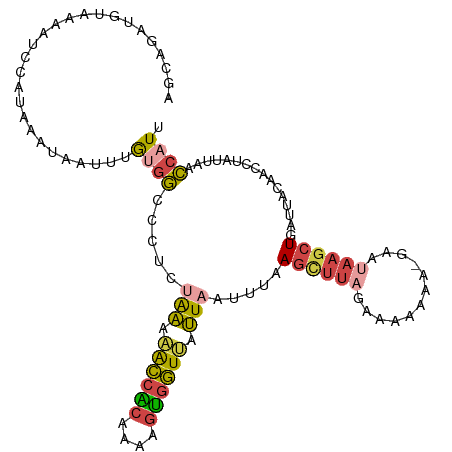
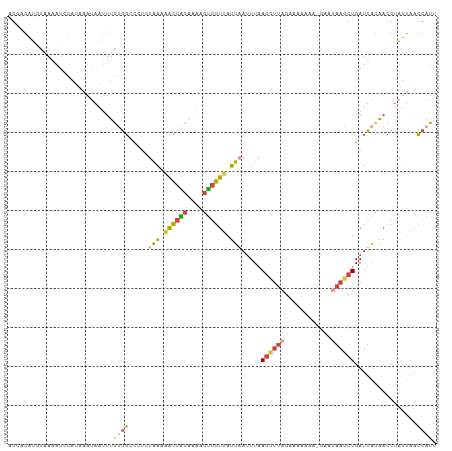
| Location | 1,845,843 – 1,845,953 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.09 |
| Shannon entropy | 0.58647 |
| G+C content | 0.30652 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -8.69 |
| Energy contribution | -8.92 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1845843 110 + 23011544 AGCAGAUUUAAAAUCCAUAAAUAAUUUGUGGCCCUCUGAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAAAGAAUAAGCUGAUUACAACCUAUUAACCAUU ..((((........((((((.....))))))...))))..((((((....))))))..(((((..((((((............)))))))))))................ ( -17.80, z-score = -1.68, R) >droEre2.scaffold_4929 1892850 109 + 26641161 GGGAGAUGCAAAAUGUAUAAAUAAUUUGCGGUCCUUUAAUAGCCACAGGAGUGGUUAUUUAAUUAAGUUUAGAAAAAAA-GCACAAGCUGAUUAUAACCUAUUAACCAUU ((..((((((((.(((....))).)))))(((.(((.(((((((((....))))))))).....)))...........(-((....))).......))).)))..))... ( -21.70, z-score = -1.29, R) >droYak2.chr2L 1819496 109 + 22324452 AGGAGUUGUAAAGCGCUUAAAUUAUUUGUGGUCUUAUGAUAACCGCGAAAGUGGUAAUUAAUUUAAGUUUAGAAAAAAA-GCAUAAGCUGAUUAUAACCUAAUGCCCAUU .((.(((((((...((((((((((......(((....))).(((((....)))))...))))))))))..........(-((....)))..))))))))).......... ( -25.50, z-score = -2.45, R) >droSec1.super_14 1792578 109 + 2068291 AGCAGAUUUAAAAUCCAUUAAUAAUUUGUGGCCCUCUGAAAACCACAAAUGUGGUUAUUAAUUUAAGCUUAGAAAAAAA-GAAUAAGCUGAUUACAACCUAUUAACCAUU .................((((((..((((((.....(((.((((((....)))))).))).....((((((........-...))))))..))))))..))))))..... ( -18.60, z-score = -1.75, R) >droSim1.chr2L 1832934 109 + 22036055 AGCUGAUUUAAAAUCCAUUAAUAAUUUGUGGCCUUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAA-GAAUAAGCUGAUUACAACCUAUUAACCAUU .................((((((..((((((.....(((.((((((....)))))).))).....((((((........-...))))))..))))))..))))))..... ( -18.20, z-score = -1.66, R) >dp4.chr4_group3 8578261 94 + 11692001 GGGAUUCGUAAUAGUUUCAAAGAAGUCAGUCCUUCCUAUAGAUCGU---UUCUAUCAAUCAUUUCAACCC-------------CAACCUGAUAGGGUUCUGAAAAGCAUU (((..................((((......))))..(((((....---.))))).............))-------------)(((((....)))))............ ( -12.90, z-score = 0.90, R) >consensus AGCAGAUGUAAAAUCCAUAAAUAAUUUGUGGCCCUCUAAAAACCACAAAAGUGGUUAUUAAUUUAAGCUUAGAAAAAAA_GAAUAAGCUGAUUACAACCUAUUAACCAUU ...........................((((.....(((.((((((....)))))).))).....((((((............))))))................)))). ( -8.69 = -8.92 + 0.23)
| Location | 1,845,843 – 1,845,953 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 69.09 |
| Shannon entropy | 0.58647 |
| G+C content | 0.30652 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -9.27 |
| Energy contribution | -9.25 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1845843 110 - 23011544 AAUGGUUAAUAGGUUGUAAUCAGCUUAUUCUUUUUUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUUUUCAGAGGGCCACAAAUUAUUUAUGGAUUUUAAAUCUGCU ..((((((((((((((....)))))))).(((........)))..........))))))..((((((((((.....))))))))))........((((.....))))... ( -21.90, z-score = -1.45, R) >droEre2.scaffold_4929 1892850 109 - 26641161 AAUGGUUAAUAGGUUAUAAUCAGCUUGUGC-UUUUUUUCUAAACUUAAUUAAAUAACCACUCCUGUGGCUAUUAAAGGACCGCAAAUUAUUUAUACAUUUUGCAUCUCCC ...((((..((((((..((..(((....))-)..)).....))))))....((((.((((....)))).))))....))))(((((............)))))....... ( -19.30, z-score = -1.22, R) >droYak2.chr2L 1819496 109 - 22324452 AAUGGGCAUUAGGUUAUAAUCAGCUUAUGC-UUUUUUUCUAAACUUAAAUUAAUUACCACUUUCGCGGUUAUCAUAAGACCACAAAUAAUUUAAGCGCUUUACAACUCCU ...(.((.(((((((((....(((....))-)................................(.((((.......)))).)..))))))))))).)............ ( -14.10, z-score = -0.21, R) >droSec1.super_14 1792578 109 - 2068291 AAUGGUUAAUAGGUUGUAAUCAGCUUAUUC-UUUUUUUCUAAGCUUAAAUUAAUAACCACAUUUGUGGUUUUCAGAGGGCCACAAAUUAUUAAUGGAUUUUAAAUCUGCU ....((((((((((((((((.((((((...-........))))))...))).))))))..(((((((((((.....))))))))))))))))))((((.....))))... ( -24.70, z-score = -2.40, R) >droSim1.chr2L 1832934 109 - 22036055 AAUGGUUAAUAGGUUGUAAUCAGCUUAUUC-UUUUUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUUUUUAGAAGGCCACAAAUUAUUAAUGGAUUUUAAAUCAGCU ..(.((((((((((((((((.((((((...-........))))))...))).))))))...((((((((((.....)))))))))).))))))).).............. ( -23.50, z-score = -2.19, R) >dp4.chr4_group3 8578261 94 - 11692001 AAUGCUUUUCAGAACCCUAUCAGGUUG-------------GGGUUGAAAUGAUUGAUAGAA---ACGAUCUAUAGGAAGGACUGACUUCUUUGAAACUAUUACGAAUCCC ......(((((((((((((......))-------------))))).....(((((......---.)))))....(((((......))))))))))).............. ( -18.70, z-score = -0.14, R) >consensus AAUGGUUAAUAGGUUGUAAUCAGCUUAUUC_UUUUUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUUUUCAGAAGGCCACAAAUUAUUUAUGGAUUUUAAAUCUCCU ..(((((..((((((......))))))...........................((((((....)))))).......)))))............................ ( -9.27 = -9.25 + -0.02)
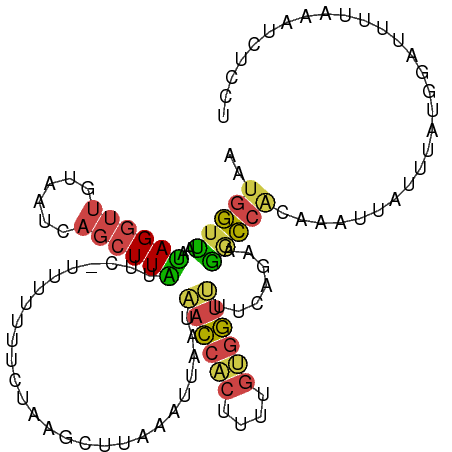
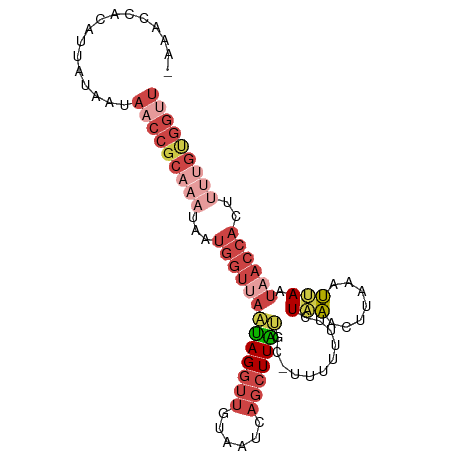
| Location | 1,845,883 – 1,845,978 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 70.15 |
| Shannon entropy | 0.57100 |
| G+C content | 0.30260 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -7.72 |
| Energy contribution | -10.32 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960271 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 1845883 95 - 23011544 -AAGCCACAUUAUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUCUUUUUUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUU -.(((((((....((((((((.......)))))....(((((((((.((((((............))))))...))).)))))))))..))))))) ( -21.70, z-score = -2.93, R) >droEre2.scaffold_4929 1892890 94 - 26641161 -AAGCCAGAUUUAAAUAACCGCAAAUAAUGGUUAAUAGGUUAUAAUCAGCUUGUGC-UUUUUUUCUAAACUUAAUUAAAUAACCACUCCUGUGGCU -.(((((.(......((((((.......))))))...((((((....(((....))-).......(((......))).)))))).....).))))) ( -16.50, z-score = -0.89, R) >droYak2.chr2L 1819536 94 - 22324452 -AAACCACAUUUAAAUAACCGCAAAUAAUGGGCAUUAGGUUAUAAUCAGCUUAUGC-UUUUUUUCUAAACUUAAAUUAAUUACCACUUUCGCGGUU -...............((((((.((...(((((((.(((((......)))))))))-(((......))).............)))..)).)))))) ( -14.60, z-score = -1.30, R) >droSec1.super_14 1792618 94 - 2068291 -AAACCACAUUAUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUC-UUUUUUUCUAAGCUUAAAUUAAUAACCACAUUUGUGGUU -.(((((((......((((((.......))))))...(((((((((.((((((...-........))))))...))).)))))).....))))))) ( -21.70, z-score = -3.46, R) >droSim1.chr2L 1832974 94 - 22036055 -AAACCAAAUUGUAGUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUUC-UUUUUUUCUAAGCUUAAAUUAAUAACCACUUUUGUGGUU -...............(((((((((...((((((((((((((....)))))))).(-((.......)))..........))))))..))))))))) ( -21.20, z-score = -2.95, R) >dp4.chr4_group3 8578299 78 - 11692001 AAAAUUGACAAGCAAUACCUCC----AAUGCUUUUCAGAACCCUAUCAGGUUGGGGUUGAAAUGAUUGAUAGAAACGAUCUA-------------- ....((((.(((((........----..))))).))))(((((((......))))))).....(((((.......)))))..-------------- ( -18.00, z-score = -1.62, R) >consensus _AAACCACAUUAUAAUAACCGCAAAUAAUGGUUAAUAGGUUGUAAUCAGCUUAUGC_UUUUUUUCUAAACUUAAAUUAAUAACCACUUUUGUGGUU ................(((((((((...(((((((((((((......)))))))...........(((.......))).))))))..))))))))) ( -7.72 = -10.32 + 2.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:30 2011