
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,925,380 – 19,925,476 |
| Length | 96 |
| Max. P | 0.573515 |

| Location | 19,925,380 – 19,925,476 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 89.76 |
| Shannon entropy | 0.21896 |
| G+C content | 0.65426 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -36.86 |
| Energy contribution | -37.78 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.573515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19925380 96 - 23011544 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUUGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUGUUGCGAAUACU ---..(((((((((((((((((((((((.....))))....)))).)))...))))))))))))..(((........)))(((((...)))))...... ( -44.20, z-score = -0.99, R) >droGri2.scaffold_15126 7183230 93 - 8399593 ------GGAUCCUCCACUGGAGCUGGCCUGGCUGGCCGCCGAGCUGCCGUUGGCAGCGGUGCCAAUGCUAUCAAGCGAUCCGCGACUGCUGCGAAUGCU ------((.....)).((.(.((.((((.....))))))).))..((.(((.((((((((((...((((....))))....)).)))))))).))))). ( -35.70, z-score = 0.43, R) >droWil1.scaffold_180708 8793858 99 + 12563649 GGAGCUGGAGCCGCCGCUGGAGCUGGCCUGACUGGCCGCUGAGCUGCCGUUUGCGGCGGUUCCAAUGCUAUCCAAUGAACCGCGACUAUUCCGUAUGCU ((((((((((((((((((((((((((((.....))))....)))).)))...))))))))))))..))..)))........(((((......)).))). ( -44.80, z-score = -2.81, R) >dp4.chr4_group4 2762713 96 + 6586962 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGACUGGCCGCAGAGCUGCCGUUUGCGGCGGUUCCAAUGCUGUCUAGAGAGCCUCGACUAUUGCGGAUUCU ---..(((((((((((((((((((((((.....))))....)))).)))...)))))))))))).(((.(((.((.....)).)))....)))...... ( -44.10, z-score = -1.74, R) >droPer1.super_10 570482 96 + 3432795 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGACUGGCCGCAGAGCUGCCGUUUGCGGCGGUUCCAAUGCUGUCUAGAGAACCUCGACUAUUGCGGAUUCU ---..(((((((((((((((((((((((.....))))....)))).)))...)))))))))))).(((.(((.((.....)).)))....)))...... ( -44.10, z-score = -2.34, R) >droSim1.chr2L 19588200 96 - 22036055 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUGUUGCGAAUACU ---..((((((((((((...(((.((((.....)))))))((((....))))))))))))))))..(((........)))(((((...)))))...... ( -45.70, z-score = -1.18, R) >droSec1.super_7 3529980 96 - 3727775 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUGUUGCGAAUACU ---..((((((((((((...(((.((((.....)))))))((((....))))))))))))))))..(((........)))(((((...)))))...... ( -45.70, z-score = -1.18, R) >droYak2.chr2R 6395023 96 - 21139217 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUAUUGCGAAUACU ---..((((((((((((...(((.((((.....)))))))((((....))))))))))))))))..(((........)))(((((...)))))...... ( -45.70, z-score = -1.44, R) >droEre2.scaffold_4845 18187768 96 + 22589142 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUAUUGCGAAUGCU ---..((((((((((((...(((.((((.....)))))))((((....))))))))))))))))..(((........)))(((((...)))))...... ( -45.70, z-score = -1.01, R) >droAna3.scaffold_12916 7290475 96 + 16180835 ---GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCAGCAGAGCUACCGUUCGCGGCGGUUCCGAUGCUGUCCAAAGAGCCUCGACUAUUGCGAAUUCU ---((((((((((((((....((((.((.....)))))).((((....)))))))))))))))((.(((........))).)).)))............ ( -42.00, z-score = -1.09, R) >consensus ___GGUGGAGCCGCCGCUGGAGCUGGCCUGGCUGGCCGCUGAGCUGCCGUUCGCGGCGGUUCCAAUGCUGUCCAAGGAGCCGCGACUAUUGCGAAUACU .....(((((((((((((((((((((((.....))))....)))).)))...)))))))))))).(((.(((...........)))....)))...... (-36.86 = -37.78 + 0.92)

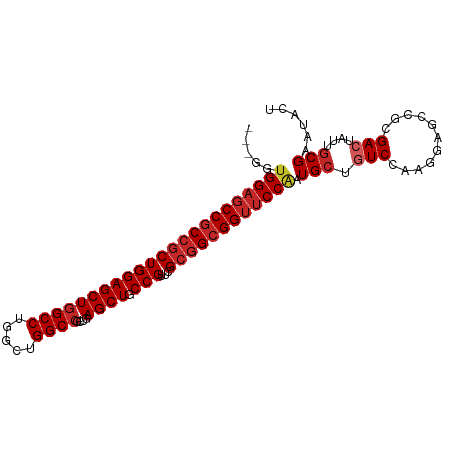
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:19 2011