
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,884,901 – 19,885,017 |
| Length | 116 |
| Max. P | 0.840545 |

| Location | 19,884,901 – 19,885,017 |
|---|---|
| Length | 116 |
| Sequences | 10 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 58.68 |
| Shannon entropy | 0.87229 |
| G+C content | 0.41689 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -5.48 |
| Energy contribution | -5.38 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
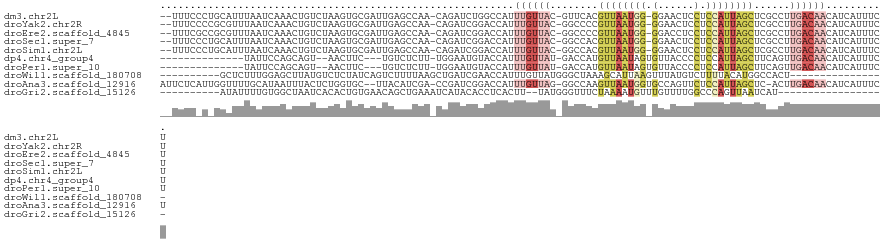
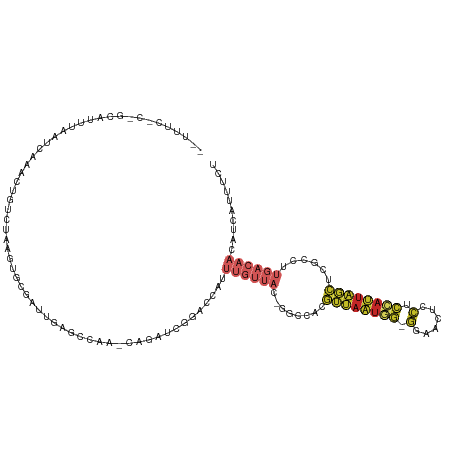
>dm3.chr2L 19884901 116 - 23011544 --UUUCCCUGCAUUUAAUCAAACUGUCUAAGUGCGAUUGAGCCAA-CAGAUCUGGCCAUUUGUUAC-GUUCACGUUAAUGG-GGAACUCCUCCAUUAGCUCGCCUUGACAACAUCAUUUCU --.....................((((.(((.((((.(((((.((-(((((......)))))))..-))))).((((((((-((.....)))))))))))))))))))))........... ( -32.20, z-score = -3.33, R) >droYak2.chr2R 6354706 116 - 21139217 --UUUCCCCGCGUUUAAUCAAACUGUCUAAGUGCGAUUGAGCCAA-CAGAUCGGACCAUUUGUUAC-GGCCCCGUUAAUGG-GGAACUCCUCCAUUAGCUCGCCUUGACAACAUCAUUUCU --.......(.((((((((..(((.....)))..)))))))))..-.(((...((....((((((.-(((...((((((((-((.....))))))))))..))).))))))..))...))) ( -33.60, z-score = -3.10, R) >droEre2.scaffold_4845 18149409 116 + 22589142 --UUUCGCCGCGUUUAAUCAAACUGUCUAAGUGCGAUUGAGCCAA-CAGAUCGGACCAUUUGUUAC-GGCCCCGUUAAUGG-GGACCUCCUCCAUUAGCUCGCCUUGACAACAUCAUUUCU --.......(.((((((((..(((.....)))..)))))))))..-.(((...((....((((((.-(((...((((((((-((.....))))))))))..))).))))))..))...))) ( -33.60, z-score = -2.67, R) >droSec1.super_7 3490107 116 - 3727775 --UUUCCCUGCAUUUAAUCAAACUGUCUAAGUGCGAUUGAGCCAA-CAGAUCGGACCAUUUGUUAC-GGCCACGUUAAUGG-GGAACUCCUCCAUUAGCUCGCCUUGACAACAUCAUUUCU --.....................((((.(((.((((.((.(((((-(((((......)))))))..-))))).((((((((-((.....)))))))))))))))))))))........... ( -32.60, z-score = -3.05, R) >droSim1.chr2L 19546395 116 - 22036055 --UUUCCCUGCAUUUAAUCAAACUGUCUAAGUGCGAUUGAGCCAA-CAGAUCGGACCAUUUGUUAC-GGCCACGUUAAUGG-GGAACUCCUCCAUUAGCUCGCCUUGACAACAUCAUUUCU --.....................((((.(((.((((.((.(((((-(((((......)))))))..-))))).((((((((-((.....)))))))))))))))))))))........... ( -32.60, z-score = -3.05, R) >dp4.chr4_group4 2706050 100 + 6586962 --------------UAUUCCAGCAGU--AACUUC---UGUCUCUU-UGGAAUGUACCAUUUGUUAU-GACCAUGUUAAUAGUGUUACCCCUCCAUUAGCUUCAGUUGACAACAUCAUUUCU --------------((((((((.((.--.((...---.))..)))-)))))))........(..((-((...(((((((.(.((((.........))))..).)))))))...))))..). ( -16.90, z-score = -1.03, R) >droPer1.super_10 513276 100 + 3432795 --------------UAUUCCAGCAGU--AACUUC---UGUCUCUU-UGGAAUGUACCAUUUGUUAU-GACCAUGUUAAUAGUGUUACCCCUCCAUUAGCUUCAGUUGACAACAUCAUUUCU --------------((((((((.((.--.((...---.))..)))-)))))))........(..((-((...(((((((.(.((((.........))))..).)))))))...))))..). ( -16.90, z-score = -1.03, R) >droWil1.scaffold_180708 8737889 95 + 12563649 ----------GCUCUUUGGAGCUUAUGUCUCUAUCAGUCUUUUAAGCUGAUCGAACCAUUUGUUAUGGGCUAAAGCAUUAAGUUUAUGUCUUUUACAUGGCCACU---------------- ----------((((....))))..(((..((.((((((.......)))))).))..)))..((((((....(((((((.......))).))))..))))))....---------------- ( -21.10, z-score = -0.99, R) >droAna3.scaffold_12916 7252219 116 + 16180835 AUUCUCAUUGGUUUUGCAUAAUUUACUCUGGUGC--UUACAUCGA-CCGAUCGGACCAUUUGUUAG-GGCCAAGUUAAUGGUGCCAGUUCUCCAUUAGCUC-ACUUGACAACAUCAUUUCU ..(((.(((((((..((((...........))))--.......))-))))).)))....(((((((-(....(((((((((.(......).))))))))).-.)))))))).......... ( -26.70, z-score = -1.29, R) >droGri2.scaffold_15126 7132371 91 - 8399593 ----------AUAUUUUGUGGCUAAUCACACUGUGAACAGCUGAAAUCAUACACCUCACUU--UAUGGGUUUCUAAAAUGUUUGUUUUGGCCCAGUUAAUCAU------------------ ----------.....(((.((((((..((((((....)))..(((((((((..........--))).)))))).........))).)))))))))........------------------ ( -15.50, z-score = 0.18, R) >consensus __UUUC_C_GCAUUUAAUCAAACUGUCUAAGUGCGAUUGAGCCAA_CAGAUCGGACCAUUUGUUAC_GGCCACGUUAAUGG_GGAACUCCUCCAUUAGCUCGCCUUGACAACAUCAUUUCU .........................................................................((((((((.(......).))))))))...................... ( -5.48 = -5.38 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:18 2011