
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,882,129 – 19,882,222 |
| Length | 93 |
| Max. P | 0.694333 |

| Location | 19,882,129 – 19,882,222 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 67.70 |
| Shannon entropy | 0.62559 |
| G+C content | 0.41621 |
| Mean single sequence MFE | -16.56 |
| Consensus MFE | -10.25 |
| Energy contribution | -10.72 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.40 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.694333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19882129 93 + 23011544 CCAUCUCAGGCCAUCGCAGCCAAAAAAAAAAAAGCCAAAUCGUUUGUGCAGUAC--G-CGUGUUGCACUUGCAGUCAUUCGUAAAUUGAAUUUACA- .....((((((.......)))............(((((.....))).))..(((--(-.(((((((....)))).))).))))...))).......- ( -16.40, z-score = 0.21, R) >droSim1.chr2L 19543605 82 + 22036055 ----------CCAUCUCGACCAGCCAAAAAAA-GCCAAAUCGCUUGUGCAGUAC--G-CGUGUUGCACUUGCAGUCAUUCGUAAAUUGAAUUUACA- ----------.......(((..((........-))......((..((((((((.--.-..))))))))..)).)))....((((((...)))))).- ( -16.80, z-score = -0.75, R) >droSec1.super_7 3487313 82 + 3727775 ----------CCAUCUCGACCAGCCAAAAAAA-GCCAAAUCGUUUGUGCAGUAC--G-CGUGUUGCACUUGCAGUCAUUCGUAAAUUGAAUUUACA- ----------.......(((..((........-))......((..((((((((.--.-..))))))))..)).)))....((((((...)))))).- ( -15.20, z-score = -0.34, R) >droYak2.chr2R 6351852 83 + 21139217 ----------CCAUCUCAGGCAGCCAAAAAAAAGCCAAAUCGCUUGUGCAGUAC--G-CGUGUUGCACUUGCAGUCAUUCGUAAAUUGAAUUUACA- ----------........(((............))).....((..((((((((.--.-..))))))))..))........((((((...)))))).- ( -17.40, z-score = -0.31, R) >droPer1.super_10 2922676 76 + 3432795 -------------------AAAAUAAAAAAGGAAGAAAAUUGUUUGUGCAACACACG-CGUGUUGCACUUUCAGUCAUUCGUAAAUUGAAUUUACA- -------------------............(((....((((...(((((((((...-.)))))))))...))))..)))((((((...)))))).- ( -20.50, z-score = -3.14, R) >droWil1.scaffold_180708 8734113 91 - 12563649 ------CUCAUUCUCUUUUUUGGCUAUCGAAAAUGGCAUGUCCUUGUGCAACACGCGUCCUUCUGCACUUGCAGUCAAUCGUAAAUUGAAUUUAAAC ------...((((...((((((.....))))))..((((......)))).....(((.....((((....)))).....))).....))))...... ( -13.80, z-score = 0.20, R) >droVir3.scaffold_12963 18835139 78 - 20206255 ----------GCAACUUUAUUU--UUGCUUGG--CCAAAUUGUAU-UGCAACGC--G-CGUGUUGCACUUGCAGCCAUUCGUAAAUUGAAUUUACA- ----------((((........--)))).(((--(.....((((.-(((((((.--.-..)))))))..))))))))...((((((...)))))).- ( -19.80, z-score = -0.96, R) >droMoj3.scaffold_6500 4179443 80 - 32352404 --------GAGCAACUUUAUUU--UCGCCUGA--GCCAAGAGU-CGAGCAGCGC--G-CGUGUUGCACUUGCAGCCAUUCGUAAAUUGAAUUUACA- --------..((((........--((((((..--....)).).-)))((((((.--.-..))))))..))))........((((((...)))))).- ( -16.30, z-score = 1.17, R) >droGri2.scaffold_15126 7129092 81 + 8399593 ----------CCAACUUUAUUCACUCGCCUGG--CCAAAUUGAACGUUGAACGC--G-CGUGUUGCACUUGCAACCAUUCGUAAAUUGAAUUUACU- ----------........(((((..(((.((.--.(((........)))..)).--)-)).(((((....)))))...........))))).....- ( -12.80, z-score = 0.32, R) >consensus __________CCAUCUCUAUCAGCUAAAAAAA_GCCAAAUCGUUUGUGCAGCAC__G_CGUGUUGCACUUGCAGUCAUUCGUAAAUUGAAUUUACA_ .........................................((..(((((((((.....)))))))))..))........((((((...)))))).. (-10.25 = -10.72 + 0.47)

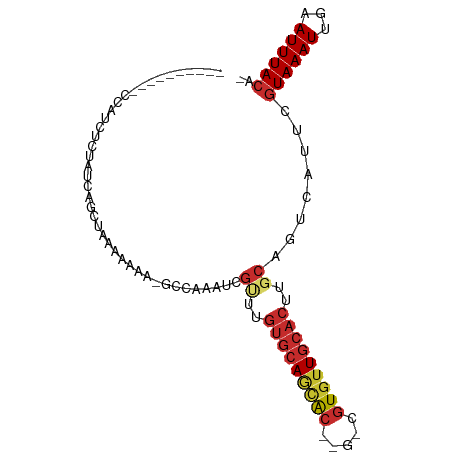

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:17 2011