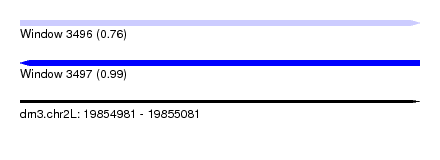
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,854,981 – 19,855,081 |
| Length | 100 |
| Max. P | 0.991508 |

| Location | 19,854,981 – 19,855,081 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 77.00 |
| Shannon entropy | 0.34882 |
| G+C content | 0.53548 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -17.95 |
| Energy contribution | -17.71 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
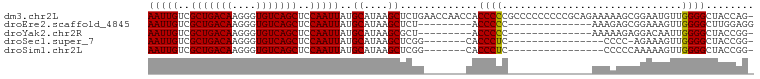
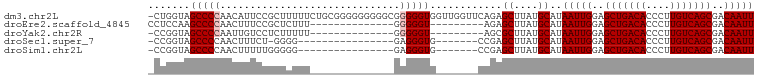
>dm3.chr2L 19854981 100 + 23011544 AAUUGUCGCUGACAAGGGUGUCAGCUCCAAUUAUGCAUAAGCUCUGAACCAACCACCCCCGCCCCCCCCCGCAGAAAAAGCGGAAUGUUGGGGCUACCAG- (((((..(((((((....)))))))..)))))..((....))..................(((((...((((.......))))......)))))......- ( -30.10, z-score = -1.71, R) >droEre2.scaffold_4845 18119783 78 - 22589142 AAUUGUCGCUGACAAGGGUGUCAGCUCCAAUUAUGCAUAAGCUCU---------ACCCCC--------------AAAGAGCGGAAAGUUGGGGCUUGGAGG (((((..(((((((....)))))))..))))).........((((---------(.((((--------------((...........))))))..))))). ( -26.70, z-score = -1.57, R) >droYak2.chr2R 6324544 77 + 21139217 AAUUGUCGCUGACAAGGGUGUCAGCUCCAAUUAUGCAUAAGCGCU---------ACCCCC--------------AAAAAGAGGACAAUUGGGGCUACCGG- (((((..(((((((....)))))))..))))).........((.(---------(.((((--------------((...........)))))).)).)).- ( -23.60, z-score = -1.39, R) >droSec1.super_7 3460366 76 + 3727775 AAUUGUCGCUGACAAGGGUGUCAGCUCCAAUUAUGCAUAAGCUCGG-------CACCCUC----------------CCCC-AGAAAGUUGGGGCUACCGG- ..(((((...)))))(((((((((((.............)))).))-------)))))..----------------((((-((....)))))).......- ( -26.12, z-score = -1.59, R) >droSim1.chr2L 19515008 77 + 22036055 AAUUGUCGCUGACAAGGGUGUCAGCUCCAAUUAUGCAUAAGCUCGG-------CACCCUC----------------CCCCCAAAAAGUUGGGGCUACCGG- ..(((((...)))))(((((((((((.............)))).))-------)))))..----------------.((((((....)))))).......- ( -26.42, z-score = -1.95, R) >consensus AAUUGUCGCUGACAAGGGUGUCAGCUCCAAUUAUGCAUAAGCUCUG_______CACCCCC______________AAAAAGCGGAAAGUUGGGGCUACCGG_ (((((..(((((((....)))))))..)))))..((....)).............((((..............................))))........ (-17.95 = -17.71 + -0.24)
| Location | 19,854,981 – 19,855,081 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.00 |
| Shannon entropy | 0.34882 |
| G+C content | 0.53548 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.47 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
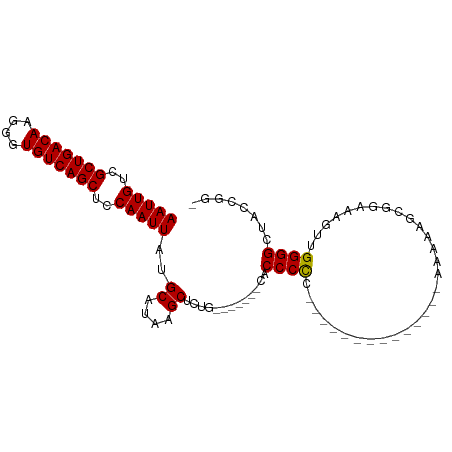
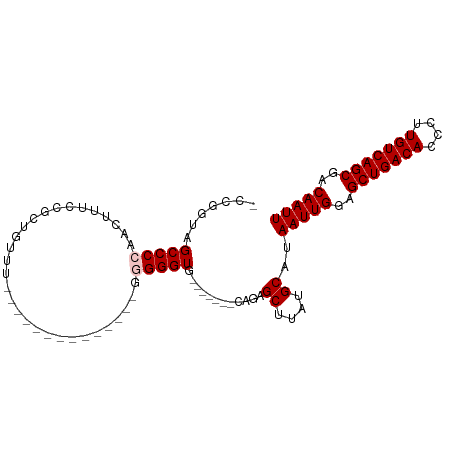
>dm3.chr2L 19854981 100 - 23011544 -CUGGUAGCCCCAACAUUCCGCUUUUUCUGCGGGGGGGGGCGGGGGUGGUUGGUUCAGAGCUUAUGCAUAAUUGGAGCUGACACCCUUGUCAGCGACAAUU -((((.....(((((.(((((((((..(.....)..)))))))))...))))).)))).((....))..(((((..(((((((....)))))))..))))) ( -41.00, z-score = -2.05, R) >droEre2.scaffold_4845 18119783 78 + 22589142 CCUCCAAGCCCCAACUUUCCGCUCUUU--------------GGGGGU---------AGAGCUUAUGCAUAAUUGGAGCUGACACCCUUGUCAGCGACAAUU ..(((((.((((((...........))--------------))))..---------...((....))....)))))(((((((....)))))))....... ( -25.60, z-score = -2.31, R) >droYak2.chr2R 6324544 77 - 21139217 -CCGGUAGCCCCAAUUGUCCUCUUUUU--------------GGGGGU---------AGCGCUUAUGCAUAAUUGGAGCUGACACCCUUGUCAGCGACAAUU -..(.((.((((((...........))--------------)))).)---------).)((....))..(((((..(((((((....)))))))..))))) ( -29.80, z-score = -3.25, R) >droSec1.super_7 3460366 76 - 3727775 -CCGGUAGCCCCAACUUUCU-GGGG----------------GAGGGUG-------CCGAGCUUAUGCAUAAUUGGAGCUGACACCCUUGUCAGCGACAAUU -.(((((.((((..(.....-)..)----------------).)).))-------))).((....))..(((((..(((((((....)))))))..))))) ( -32.70, z-score = -3.18, R) >droSim1.chr2L 19515008 77 - 22036055 -CCGGUAGCCCCAACUUUUUGGGGG----------------GAGGGUG-------CCGAGCUUAUGCAUAAUUGGAGCUGACACCCUUGUCAGCGACAAUU -.(((((.((((..((....))..)----------------).)).))-------))).((....))..(((((..(((((((....)))))))..))))) ( -33.60, z-score = -3.38, R) >consensus _CCGGUAGCCCCAACUUUCCGCUGUUU______________GGGGGUG_______CAGAGCUUAUGCAUAAUUGGAGCUGACACCCUUGUCAGCGACAAUU .......(((((..............................)))))............((....))..(((((..(((((((....)))))))..))))) (-21.07 = -21.47 + 0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:16 2011