
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,834,335 – 19,834,393 |
| Length | 58 |
| Max. P | 0.861144 |

| Location | 19,834,335 – 19,834,393 |
|---|---|
| Length | 58 |
| Sequences | 7 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 69.33 |
| Shannon entropy | 0.62353 |
| G+C content | 0.42137 |
| Mean single sequence MFE | -14.05 |
| Consensus MFE | -5.81 |
| Energy contribution | -6.96 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
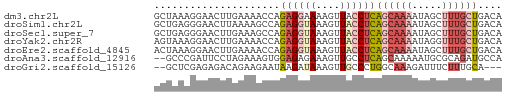
>dm3.chr2L 19834335 58 + 23011544 GCUAAAGGAACUUGAAAACCAGAGGAAAAGUUACCUCAGCAAAAUAGCUUUGCUGACA ......(((((((..............))))).))((((((((.....)))))))).. ( -12.84, z-score = -1.62, R) >droSim1.chr2L 19493991 58 + 22036055 GCUGAGGGAACUUAAAAGCCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACA (((((((.(((((....(((...))).))))).)))))))....((((...))))... ( -18.90, z-score = -2.67, R) >droSec1.super_7 3439754 58 + 3727775 GCUGAGGGAACUUGAAAGCCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACA (((((((.(((((....(((...))).))))).)))))))....((((...))))... ( -20.00, z-score = -2.85, R) >droYak2.chr2R 6303298 58 + 21139217 AGUAAAGGAACUUGAAAACCAGAGGUAAAGUUACCUCAGCAAAAUAGGUUUGCUGACA ......(((((((....(((...))).))))).))((((((((.....)))))))).. ( -14.30, z-score = -2.01, R) >droEre2.scaffold_4845 18099215 58 - 22589142 ACUAAAGGAACUUGAAAACCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACA ......(((((((....(((...))).))))).))((((((((.....)))))))).. ( -14.50, z-score = -2.37, R) >droAna3.scaffold_12916 7201163 56 - 16180835 --GCCCGAUUCCUAGAAAGUGGAGAGAAAGUUGCCUCAGCAAAAAUGCGCAGAUGCCA --((.....(((........)))(((........))).(((....)))))........ ( -8.20, z-score = 0.77, R) >droGri2.scaffold_15126 7073010 53 + 8399593 --GCUCGAGAGACAGAAGAAUAAGAUAAAGUUGCCCUGGCAAAGAUUUCUUUGCA--- --((((....)).........((((.....((((....)))).....)))).)).--- ( -9.60, z-score = -0.29, R) >consensus GCUAAAGGAACUUGAAAACCAGAGGUAAAGUUACCUCAGCAAAAUAGCUUUGCUGACA .....................((((((....))))))((((((.....)))))).... ( -5.81 = -6.96 + 1.14)

| Location | 19,834,335 – 19,834,393 |
|---|---|
| Length | 58 |
| Sequences | 7 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 69.33 |
| Shannon entropy | 0.62353 |
| G+C content | 0.42137 |
| Mean single sequence MFE | -12.25 |
| Consensus MFE | -6.12 |
| Energy contribution | -5.80 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
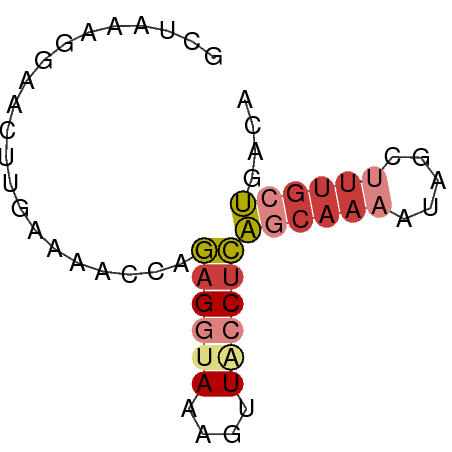
>dm3.chr2L 19834335 58 - 23011544 UGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUUCCUCUGGUUUUCAAGUUCCUUUAGC ..(((((((((...)))))))))((.(((((..((...)).....)))))))...... ( -12.10, z-score = -0.89, R) >droSim1.chr2L 19493991 58 - 22036055 UGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGCUUUUAAGUUCCCUCAGC ....(((...))).....(((((((.(((((..............))))).))))))) ( -14.64, z-score = -1.31, R) >droSec1.super_7 3439754 58 - 3727775 UGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGCUUUCAAGUUCCCUCAGC ....(((...))).....(((((((.(((((..............))))).))))))) ( -13.94, z-score = -1.04, R) >droYak2.chr2R 6303298 58 - 21139217 UGUCAGCAAACCUAUUUUGCUGAGGUAACUUUACCUCUGGUUUUCAAGUUCCUUUACU ..((((((((.....))))))))((.(((((.(((...)))....)))))))...... ( -14.20, z-score = -2.58, R) >droEre2.scaffold_4845 18099215 58 + 22589142 UGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGUUUUCAAGUUCCUUUAGU ..(((((((((...)))))))))((.(((((.(((...)))....)))))))...... ( -14.00, z-score = -1.75, R) >droAna3.scaffold_12916 7201163 56 + 16180835 UGGCAUCUGCGCAUUUUUGCUGAGGCAACUUUCUCUCCACUUUCUAGGAAUCGGGC-- ..((....))(((....))).(((....)))....(((........))).......-- ( -8.30, z-score = 1.41, R) >droGri2.scaffold_15126 7073010 53 - 8399593 ---UGCAAAGAAAUCUUUGCCAGGGCAACUUUAUCUUAUUCUUCUGUCUCUCGAGC-- ---.((((((....)))))).(((....))).........................-- ( -8.60, z-score = -0.24, R) >consensus UGUCAGCAAAGCUAUUUUGCUGAGGUAACUUUACCUCUGGUUUUCAAGUUCCUUUAGC ....((((((.....))))))((((........))))..................... ( -6.12 = -5.80 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:12 2011