| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,825,859 – 19,825,953 |
| Length | 94 |
| Max. P | 0.896023 |

| Location | 19,825,859 – 19,825,953 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.09 |
| Shannon entropy | 0.51821 |
| G+C content | 0.50125 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -8.66 |
| Energy contribution | -9.56 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
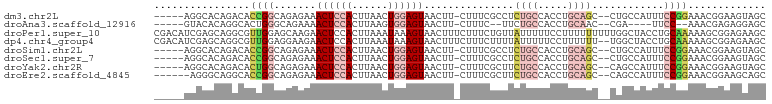
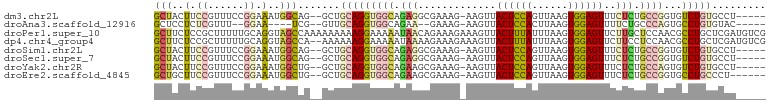
>dm3.chr2L 19825859 94 + 23011544 -----AGGCACAGACACCGGCAGAGAAACUCCACUUAACUGGAGUAACUU-CUUUCGCCUCUGCCACCUGCAGC--CUGCCAUUUCCGGAAACGGAAGUAGC -----.((((........(((((((((((((((......))))))...))-)))).))).((((.....)))).--.))))(((((((....)))))))... ( -37.30, z-score = -4.39, R) >droAna3.scaffold_12916 7192754 86 - 16180835 -----GUACACAGGCACUGGGCAGAAAACUCCACUUAAGUGGAGUAACUU-CUUUC--UUCUGCCACCUGCAAC--CGA----UUCC--AAACGAGAGGAGC -----........(((...(((((((.(((((((....))))))).....-.....--)))))))...)))...--...----((((--........)))). ( -25.70, z-score = -1.99, R) >droPer1.super_10 420799 102 - 3432795 CGACAUCGAGCAGGCGUUGGAGCAAGAACUCCACUUAAAUAAAGUAACUUUCUUUCUGUUAUUUUUCCUUUUUUUUUGGCUACCUGCAAAAAGCGGAGAAGC ........((((((....((((......))))((((.....)))).........)))))).(((((((.((((((...((.....)))))))).))))))). ( -19.40, z-score = 0.73, R) >dp4.chr4_group4 2596166 100 - 6586962 CGACAUCGAGCAGGCGUUGGAGGAAGAACUCCACUUAAAUAAAGUAACUUUCUUUCUUUUAUUUUUCCUUUUUU--UGGCUACCUGCAAAAAGCGGAGAAGC ((((..(.....)..))))(((((((((....((((.....))))....)))))))))...(((((((.(((((--((.(.....)))))))).))))))). ( -22.90, z-score = -0.47, R) >droSim1.chr2L 19485460 94 + 22036055 -----AGGCACAGACACCGGCAGAGAAACUCCACUUAACUGGAGUAACUU-CUUUCGCCUCUGCCACCUGCAGC--CUGCCAUUUCCGGAAACGGAAGUAGC -----.((((........(((((((((((((((......))))))...))-)))).))).((((.....)))).--.))))(((((((....)))))))... ( -37.30, z-score = -4.39, R) >droSec1.super_7 3431234 94 + 3727775 -----AGGCACAGACACCGGCAGAGAAACUCCACUUAACUGGAGUAACUU-CUUUCGCCUCUGCCACCUGCAGC--CUGCCAUUUCCGGAAACGGAAGUAGC -----.((((........(((((((((((((((......))))))...))-)))).))).((((.....)))).--.))))(((((((....)))))))... ( -37.30, z-score = -4.39, R) >droYak2.chr2R 6294937 94 + 21139217 -----AGGCACAGACACUGGCAGAGAAACUCCACUUAACUGGAGUAACUU-CUUUCGCUUCUGCCACCUGCAGC--CAGCCAUUUCCGGAAACGGAAGUAGC -----.(((.(((....((((((((((((((((......)))))).....-..)))...))))))).)))..))--).((.(((((((....))))))).)) ( -36.31, z-score = -4.09, R) >droEre2.scaffold_4845 18090665 93 - 22589142 ------AGGGCAGGCACCGGCAGAGAAACUCCACUUAACUGGAGUAACUU-CUUUCGCUUCUGCCACCUGCAGC--CAGCCAUUUCCGGAAACGGAAGCAGC ------..((((((....(((((((((((((((......))))))...))-)))).)))))))))..((((.(.--...)...(((((....))))))))). ( -36.00, z-score = -2.92, R) >consensus _____AGGCACAGACACCGGCAGAGAAACUCCACUUAACUGGAGUAACUU_CUUUCGCUUCUGCCACCUGCAGC__CGGCCAUUUCCGGAAACGGAAGUAGC ................((((.......((((((......))))))...............((((.....))))............))))............. ( -8.66 = -9.56 + 0.91)
| Location | 19,825,859 – 19,825,953 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 73.09 |
| Shannon entropy | 0.51821 |
| G+C content | 0.50125 |
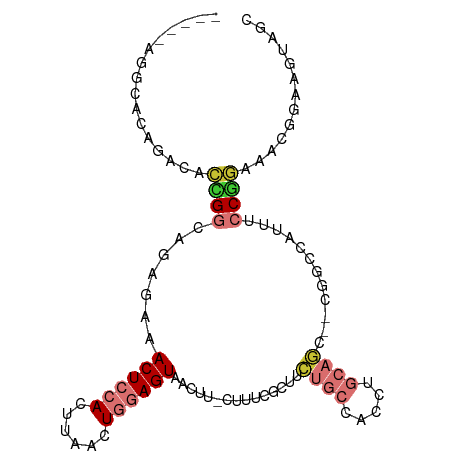
| Mean single sequence MFE | -34.71 |
| Consensus MFE | -11.84 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19825859 94 - 23011544 GCUACUUCCGUUUCCGGAAAUGGCAG--GCUGCAGGUGGCAGAGGCGAAAG-AAGUUACUCCAGUUAAGUGGAGUUUCUCUGCCGGUGUCUGUGCCU----- ((((.(((((....))))).))))((--((.(((((((((((((((....)-.....((((((......)))))).)))))))))...)))))))))----- ( -42.60, z-score = -3.83, R) >droAna3.scaffold_12916 7192754 86 + 16180835 GCUCCUCUCGUUU--GGAA----UCG--GUUGCAGGUGGCAGAA--GAAAG-AAGUUACUCCACUUAAGUGGAGUUUUCUGCCCAGUGCCUGUGUAC----- ..(((........--))).----...--...(((((((((((((--(....-.....(((((((....)))))))))))))))....))))))....----- ( -30.70, z-score = -2.58, R) >droPer1.super_10 420799 102 + 3432795 GCUUCUCCGCUUUUUGCAGGUAGCCAAAAAAAAAGGAAAAAUAACAGAAAGAAAGUUACUUUAUUUAAGUGGAGUUCUUGCUCCAACGCCUGCUCGAUGUCG ((......)).....((((((..((.........))..................(((((((.....))))(((((....))))))))))))))......... ( -19.70, z-score = 0.29, R) >dp4.chr4_group4 2596166 100 + 6586962 GCUUCUCCGCUUUUUGCAGGUAGCCA--AAAAAAGGAAAAAUAAAAGAAAGAAAGUUACUUUAUUUAAGUGGAGUUCUUCCUCCAACGCCUGCUCGAUGUCG ((......)).....((((((..((.--......)).........((.(((((..((((((.....))))))..))))).)).....))))))......... ( -20.10, z-score = -0.29, R) >droSim1.chr2L 19485460 94 - 22036055 GCUACUUCCGUUUCCGGAAAUGGCAG--GCUGCAGGUGGCAGAGGCGAAAG-AAGUUACUCCAGUUAAGUGGAGUUUCUCUGCCGGUGUCUGUGCCU----- ((((.(((((....))))).))))((--((.(((((((((((((((....)-.....((((((......)))))).)))))))))...)))))))))----- ( -42.60, z-score = -3.83, R) >droSec1.super_7 3431234 94 - 3727775 GCUACUUCCGUUUCCGGAAAUGGCAG--GCUGCAGGUGGCAGAGGCGAAAG-AAGUUACUCCAGUUAAGUGGAGUUUCUCUGCCGGUGUCUGUGCCU----- ((((.(((((....))))).))))((--((.(((((((((((((((....)-.....((((((......)))))).)))))))))...)))))))))----- ( -42.60, z-score = -3.83, R) >droYak2.chr2R 6294937 94 - 21139217 GCUACUUCCGUUUCCGGAAAUGGCUG--GCUGCAGGUGGCAGAAGCGAAAG-AAGUUACUCCAGUUAAGUGGAGUUUCUCUGCCAGUGUCUGUGCCU----- ((((.(((((....))))).)))).(--((.((((((((((((..(....)-..(..((((((......))))))..))))))))...)))))))).----- ( -40.00, z-score = -3.64, R) >droEre2.scaffold_4845 18090665 93 + 22589142 GCUGCUUCCGUUUCCGGAAAUGGCUG--GCUGCAGGUGGCAGAAGCGAAAG-AAGUUACUCCAGUUAAGUGGAGUUUCUCUGCCGGUGCCUGCCCU------ (((..(((((....)))))..))).(--(..((((((((((((..(....)-..(..((((((......))))))..)))))))...)))))))).------ ( -39.40, z-score = -2.60, R) >consensus GCUACUUCCGUUUCCGGAAAUGGCAG__GCUGCAGGUGGCAGAAGCGAAAG_AAGUUACUCCAGUUAAGUGGAGUUUCUCUGCCAGUGCCUGUGCCU_____ (((....((......))....))).......(((((((((.(((.............((((((......))))))..))).))))...)))))......... (-11.84 = -12.44 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:10 2011