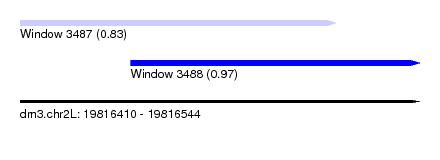
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,816,410 – 19,816,544 |
| Length | 134 |
| Max. P | 0.967604 |

| Location | 19,816,410 – 19,816,516 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Shannon entropy | 0.39117 |
| G+C content | 0.36416 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.04 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.828519 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 19816410 106 + 23011544 GUCACUCUGGUUUCUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACACAAUUUUUA--UUGAGGC----- ....(((..(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-.............--..)))..----- ( -22.80, z-score = -1.28, R) >droSim1.chr2L 19476382 105 + 22036055 -UCACUCUGGUUUCUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACUCAAUUUUUA--UUGAGGC----- -........(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-..((((((....)--)))))..----- ( -25.50, z-score = -2.35, R) >droSec1.super_7 3422959 106 + 3727775 GUCACUCUGGUUUCUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGAAACACUCUAAUGAGUCUA-AACUCAAUUUUUA--UUGAGGC----- .........(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-..((((((....)--)))))..----- ( -26.00, z-score = -2.64, R) >droYak2.chr2R 6285830 106 + 21139217 GUCACUCUGGUUUCUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACUCAAUUUUUA--UUAGGGC----- ....((((((((((((((((..---...((...((((((((.(.......).)-))))))).))))))))))))).......(((((...-.))))).......--.))))).----- ( -24.40, z-score = -1.92, R) >droEre2.scaffold_4845 4510125 106 + 22589142 GUCACUCUGGUUUCUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACUCAAUUUUUA--UUAAGGC----- (((......(((((((((((..---...((...((((((((.(.......).)-))))))).))))))))))))).......(((((...-.))))).......--....)))----- ( -22.20, z-score = -1.52, R) >droAna3.scaffold_13340 22982763 112 + 23697760 ------GAAAACUCUAUUUAAUGCAUAUUUAAUAACAUUUUAAAUGCCUAAUAUAAAAACUAAAAGCAAAUUAAGGCUCAAAUGUGUAUAUAAAUGAGUUGGCGCGAAAGAGCUGUGU ------...(((((.(((((((((((((((.......(((((.((.....)).)))))......(((........))).)))))))))).))))))))))(((.(....).))).... ( -22.20, z-score = -1.69, R) >dp4.chr4_group4 4127631 107 + 6586962 GUUACUCUGGCUUCUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUA-AAAUGAAUUUUUUC-GUAGUGC----- ..((((...(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-..(((((....)))-)))))).----- ( -23.90, z-score = -1.12, R) >droPer1.super_10 3027263 105 + 3432795 UUCACUCUGGUUUUUCUUCAAU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAAGAACACUCUAAUGAGUCUA-AAAU-AAUUUUUA--CCAGUGC----- ....(.((((((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-....-........--.))).).----- ( -20.20, z-score = -1.48, R) >droWil1.scaffold_180708 410365 106 - 12563649 AUCACUCUGGCUUCUCUUCAGU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUA-AAUUGAAUUUUUU--GAUGAGC----- ....(((..(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-.............--...))).----- ( -23.50, z-score = -0.35, R) >droGri2.scaffold_8087 3221 105 + 4350 UUCACUCUGGCUUCUCUUCAGU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAGCAUUCUAAUGAGUCUA-AACUCAAUUUUUU---AAGUGC----- ..((((...(((((((((((..---...((...((((((((.(.......).)-))))))).))))))))))))).......(((((...-.))))).......---.)))).----- ( -25.50, z-score = -1.96, R) >droVir3.scaffold_12963 1451896 105 + 20206255 UUCACUCUGGCUUCUCUUCAGU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUA-AAUUGAAUUUUUC---GUGUGC----- ..(((.(..(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-.............---).))).----- ( -24.30, z-score = -1.09, R) >droMoj3.scaffold_6500 3962154 106 - 32352404 UUCACUCUGGCUUCUCUUCAGU---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUA-AAAUGAAUUUUUU--GUAGUGC----- ..(((((..(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-.............--).)))).----- ( -24.40, z-score = -1.03, R) >anoGam1.chr3R 16149697 105 + 53272125 GGCACUCUGAUCUCUCUUCAAC---UGUCGAAUAAAUCUUUCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGGAUACUCUAAUGAGUCUA-UAGUGAAUUUUUG--UCCGUC------ ..((((...(((((((((((..---...((...((((((((.(.......).)-))))))).)))))))))))))((((....))))...-.))))........--......------ ( -22.80, z-score = -0.54, R) >consensus GUCACUCUGGUUUCUCUUCAAU___UGUCGAAUAAAUCUUUCGCCUUUUACUA_AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA_AACUCAAUUUUUA__UUAGUGC_____ .........(((((((((((........((((.......)))).(((.......))).......)))))))))))((((....))))............................... (-16.79 = -16.04 + -0.75)

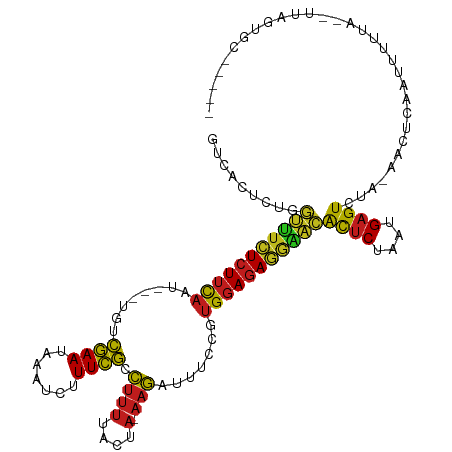
| Location | 19,816,447 – 19,816,544 |
|---|---|
| Length | 97 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.56 |
| Shannon entropy | 0.53284 |
| G+C content | 0.40259 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.68 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967604 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 19816447 97 + 23011544 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACACAAUUUUUA-UUGAGGCCUG-AUAACUUAUGCUA-UCGGGCCAAUGA----------- .............-.((((((...(((((.......)))))..)))))).-....((((....)-))).((((((-(((.(....).))-))))))).....----------- ( -24.90, z-score = -2.34, R) >droSim1.chr2L 19476418 97 + 22036055 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACUCAAUUUUUA-UUGAGGCCUG-AUAACUUAUGUUA-UCGGGCCAAUUA----------- .............-.((((((...(((((.......)))))..)))))).-...(((((....)-))))((((((-(((((....))))-))))))).....----------- ( -30.30, z-score = -4.20, R) >droSec1.super_7 3422996 97 + 3727775 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGAAACACUCUAAUGAGUCUA-AACUCAAUUUUUA-UUGAGGCCUG-AUAACUUAUGUUA-UCGGGCCAAUUA----------- .............-.((((((...(((((.......)))))..)))))).-...(((((....)-))))((((((-(((((....))))-))))))).....----------- ( -30.30, z-score = -4.64, R) >droYak2.chr2R 6285867 97 + 21139217 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACUCAAUUUUUA-UUAGGGCCUG-ACAGCUUAUGUUA-UUGGGCCAAUAG----------- .......(((.((-((((((((....))(((....((((....))))...-..)))))))))))-.)))((((..-(.(((....))).-)..)))).....----------- ( -23.20, z-score = -1.22, R) >droEre2.scaffold_4845 4510162 97 + 22589142 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA-AACUCAAUUUUUA-UUAAGGCCUG-ACAGCUUAUGUUA-UCGGGCCCAUUA----------- .......(((.((-((((((((....))(((....((((....))))...-..)))))))))))-.)))((((((-(.(((....))).-))))))).....----------- ( -25.20, z-score = -2.17, R) >droAna3.scaffold_13340 22982797 106 + 23697760 UAAAUGCCUAAUAUAAAAACUAAAAGCAAAUUAAGGCUCAAAUGUGUAUAUAAAUGAGUUGGCGCGAAAGAGCUGUGUUUCGUAAUAUAUCUUAGCAUAUAAUCUA------- ...((((...(((((...((....(((........))).....)).)))))..(((((.((((.(....).))))...)))))...........))))........------- ( -16.00, z-score = 0.64, R) >dp4.chr4_group4 4127668 98 + 6586962 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUA-AAAUGAAUUUUUUCGUAGUGCCCG-GCGGCUUAGGUUG-CUGGGCCAAUGU----------- ..(((((((.(((-..(....)..))).)))))))((((....))))...-..(((((....))))).(.(((((-(((((....))))-))))))).....----------- ( -35.00, z-score = -2.53, R) >droPer1.super_10 3027300 97 + 3432795 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAAGAACACUCUAAUGAGUCUA-AAAU-AAUUUUUA-CCAGUGCCUG-GUCUGUUCUACGGACCGGGUCAAUGA----------- .............-.((((((...(((((.......)))))..)))))).-....-........-.((.((((((-(((((.....))))))))).)).)).----------- ( -26.50, z-score = -1.99, R) >droWil1.scaffold_180708 410402 97 - 12563649 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGGGCACUCUAAUGAGUCUA-AAUUGAAUUUUUU-GAUGAGCCUA-GCGACUUUUGUUG-CCGGGCCAAUAA----------- ..(((((((.(((-..(....)..))).)))))))((((....))))...-.............-..((.(((..-(((((....))))-)..)))))....----------- ( -29.10, z-score = -2.13, R) >droGri2.scaffold_8087 3258 96 + 4350 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAGCAUUCUAAUGAGUCUA-AACUCAAUUUUUU--AAGUGCCUG-GCGGCAUUUGCAG-CCGGGCCAAUUU----------- ..(((((((.(((-..(....)..)))))))).)).......(((((...-.))))).......--..(.(((((-((.((....)).)-))))))).....----------- ( -28.50, z-score = -1.57, R) >droVir3.scaffold_12963 1451933 96 + 20206255 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUA-AAUUGAAUUUUUC-G-UGUGCCUG-GUGACUUUUGUUG-CCGGGCCAAUUG----------- ..(((((((.(((-..(....)..)))))))).))((((....))))...-.............-.-((.(((((-(..((....))..-))))))))....----------- ( -28.10, z-score = -2.19, R) >droMoj3.scaffold_6500 3962191 97 - 32352404 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUA-AAAUGAAUUUUUU-GUAGUGCCUA-GCAGCUUCGGUUG-CCGGGCCAAUUG----------- ........(((.(-(((((((((.......))((.((((....)))))).-....)))))))).-)))(.(((..-(((((....))))-)..)))).....----------- ( -27.70, z-score = -1.38, R) >anoGam1.chr3R 16149734 107 + 53272125 UCGCCUUUUACUA-AAGAUUUCCGUGGAGAGGGAUACUCUAAUGAGUCUA-UAGUGAAUUUUUG--UCCGUCUCG-AUUCCGUAAGGAG-UCGAGCCCUAAACUUCAAUACAA ......(((((((-.((((((...(((((.......)))))..)))))).-)))))))......--...(.((((-(((((....))))-))))))................. ( -31.70, z-score = -2.60, R) >consensus UCGCCUUUUACUA_AAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUA_AACUCAAUUUUUA_GUAGGGCCUG_GUAACUUAUGUUG_CCGGGCCAAUUA___________ ...............((((((...(((((.......)))))..)))))).....................(((((.(((((....)))).))))))................. (-12.67 = -12.68 + 0.02)
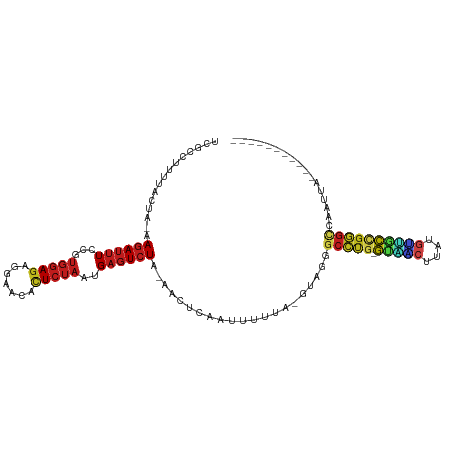
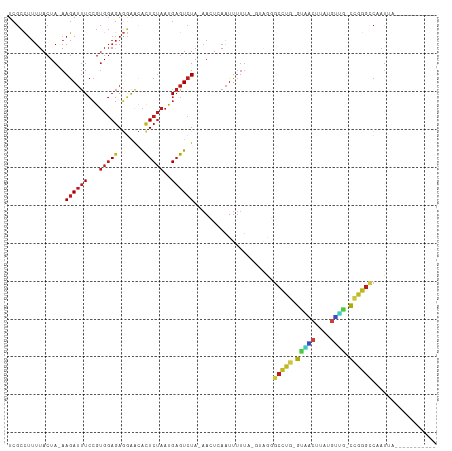
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:09 2011