| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,811,988 – 19,812,073 |
| Length | 85 |
| Max. P | 0.890004 |

| Location | 19,811,988 – 19,812,073 |
|---|---|
| Length | 85 |
| Sequences | 12 |
| Columns | 85 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Shannon entropy | 0.15534 |
| G+C content | 0.37264 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.32 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.890004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19811988 85 - 23011544 ACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -19.90, z-score = -1.95, R) >droSim1.chr2L 19471131 85 - 22036055 ACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -19.90, z-score = -1.95, R) >droSec1.super_7 3417380 85 - 3727775 ACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -19.90, z-score = -1.95, R) >droYak2.chr2R 6274650 85 - 21139217 ACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -19.90, z-score = -1.95, R) >droEre2.scaffold_4845 18076384 82 + 22589142 AUUCUGG---ACAUUCAAUUGUCGAAAAAAUCUUUCGCCUUUUAGUAAAGAUUUCCGUGGGGAGGAACACUCUAAUGAGUAGAAU (((((((---(((......))))....((((((((.((......)))))))))).(((..((((.....)))).)))..)))))) ( -16.70, z-score = -0.27, R) >droAna3.scaffold_12916 7183398 85 + 16180835 ACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGGACACACUAAUGAGUCUAGA ((((((((..(((((((.....((...((((((((.(.......).)))))))).)))))))))..)).)).....))))..... ( -18.40, z-score = -0.45, R) >dp4.chr4_group4 2582433 85 + 6586962 ACUCUGGUUUUUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAAGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -17.80, z-score = -1.66, R) >droPer1.super_10 405444 85 + 3432795 ACUCUGGUUUUUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAAGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -17.80, z-score = -1.66, R) >droMoj3.scaffold_6500 4054600 85 + 32352404 ACUCUGGCUUAUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUGAA .((((((...(((((.(((.(.((((.......)))).)....))).)))))..))..))))...((.((((....))))))... ( -17.30, z-score = 0.15, R) >droVir3.scaffold_12963 18754271 85 + 20206255 ACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -22.30, z-score = -1.81, R) >droGri2.scaffold_15126 7053371 85 - 8399593 ACUCUGGCUUCUCUUCAGUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAGCACUCUAAUGAGUCUAAA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -22.30, z-score = -1.81, R) >anoGam1.chr3R 16148468 85 - 53272125 ACUCUGAUCUCUCUUCAACUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGGAUACUCUAAUGAGUCUAUA ......(((((((((((.....((...((((((((.(.......).)))))))).)))))))))))))((((....))))..... ( -19.90, z-score = -1.72, R) >consensus ACUCUGGUUUCUCUUCAAUUGUCGAAUAAAUCUUUCGCCUUUUACUAAAGAUUUCCGUGGAGAGGAACACUCUAAUGAGUCUAAA ......(((((((((((.....((((.......)))).((((....)))).......)))))))))))((((....))))..... (-18.63 = -18.32 + -0.31)

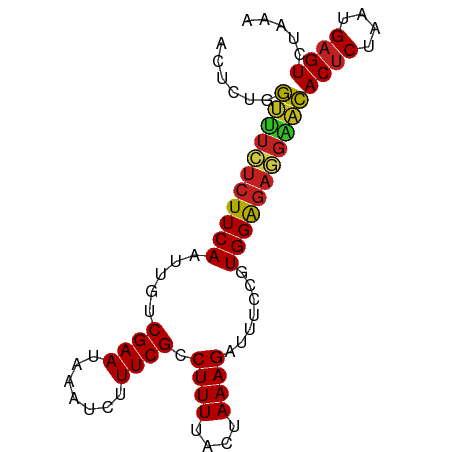
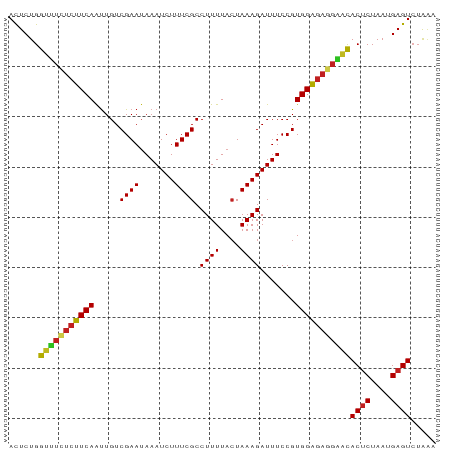
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:53:02 2011