| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,831,670 – 1,831,778 |
| Length | 108 |
| Max. P | 0.995560 |

| Location | 1,831,670 – 1,831,778 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.34 |
| Shannon entropy | 0.61341 |
| G+C content | 0.32389 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -8.62 |
| Energy contribution | -9.27 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
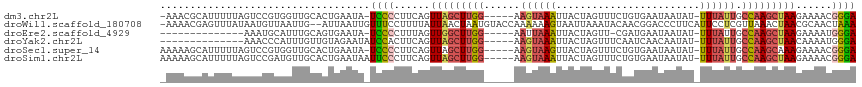
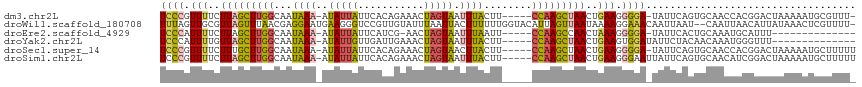
>dm3.chr2L 1831670 108 + 23011544 -AAACGCAUUUUUAGUCCGUGGUUGCACUGAAUA-UCCCCUUCAGUUAGCUUGG-----AAGUAAAUUACUAGUUUCUGUGAAUAAUAU-UUUAUUGCCAAGCUAAGAAAACGGGA -..............(((((..((..((((((..-.....))))))((((((((-----.((((((((((........)))).......-)))))).)))))))).))..))))). ( -25.61, z-score = -1.63, R) >droWil1.scaffold_180708 721781 113 - 12563649 -AAAACGAGUUUAUAAUGUUAAUUG--AUUAAUUGUUUCCUUUUAUUAACUAAUGUACCAAAAAAGUAAUUAAAUACAACGGACCCUUCAUUCCUCGUUAAACUAACGCAACUAAA -..((((((........(((..(((--.(((((((....(((((....((....)).....))))))))))))...)))..))).........))))))................. ( -9.63, z-score = 0.73, R) >droEre2.scaffold_4929 1878584 94 + 26641161 --------------AAAUGCAUUUGCAGUGAAUA-UCCCCUUUAGUUGGCUUGG-----AAUUAAAUUACUAGUU-CGAUGAAUAAUAU-UUUAUUGCCAAGCUAAGAAAAUGGGA --------------...(((....))).......-((((.(((..(((((((((-----(((((......)))))-(((((((......-))))))))))))))))..))).)))) ( -24.70, z-score = -2.82, R) >droYak2.chr2L 1804987 96 + 22324452 --------------AAACCCAUUUGUUGUAGAAUAUCCACUUCAGUUAGCUUGG-----AAGUAAAUUACUAGUUUCAAUCAACAAUAU-UUUAUUGCCAAGCUAACAAAAUGGGA --------------...(((((((......(((.......))).((((((((((-----.((((((.((...(((......)))..)).-)))))).)))))))))).))))))). ( -27.80, z-score = -4.70, R) >droSec1.super_14 1778365 109 + 2068291 AAAAAGCAUUUUUAGUCCGUGGUUGCACUGAAUA-UCCCCUUCAGUUAGCUUGG-----AAGUAAGUUACUAGUUUCUGUGAAUAAUAU-UUUAUUGCCAAGCAAAGAAAACGGGA ...............(((((..((..((((((..-.....))))))..((((((-----.((((((((((........)))).......-)))))).))))))...))..))))). ( -23.31, z-score = -0.79, R) >droSim1.chr2L 1818710 110 + 22036055 AAAAAGCAUUUUUAGUCCGAUGUUGCACUGAAUAAUUCCCUUCAGUUAGCUUGG-----AAGUAAAUUACUAGUUUCUGUGAAUAAUAU-UUUAUUGCCAAGCUAAGAAAACGGGA ...............((((.......((((((........))))))((((((((-----.((((((((((........)))).......-)))))).))))))))......)))). ( -24.01, z-score = -1.51, R) >consensus _AAAAGCAUUUUUAAACCGUAGUUGCACUGAAUA_UCCCCUUCAGUUAGCUUGG_____AAGUAAAUUACUAGUUUCUAUGAAUAAUAU_UUUAUUGCCAAGCUAAGAAAACGGGA ...................................((((......(((((((((......(((.....)))..........(((((.....))))).)))))))))......)))) ( -8.62 = -9.27 + 0.64)
| Location | 1,831,670 – 1,831,778 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.34 |
| Shannon entropy | 0.61341 |
| G+C content | 0.32389 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -11.66 |
| Energy contribution | -11.87 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
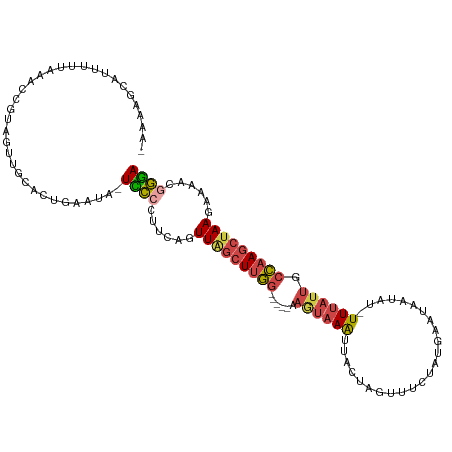
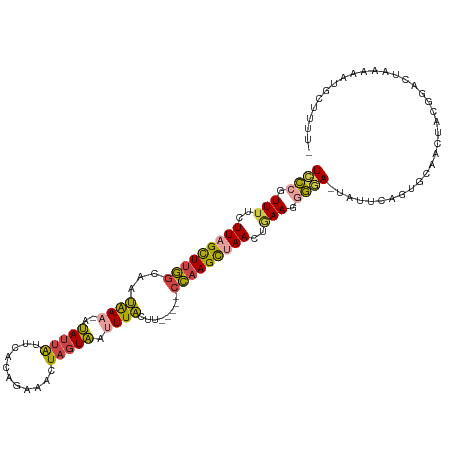
>dm3.chr2L 1831670 108 - 23011544 UCCCGUUUUCUUAGCUUGGCAAUAAA-AUAUUAUUCACAGAAACUAGUAAUUUACUU-----CCAAGCUAACUGAAGGGGA-UAUUCAGUGCAACCACGGACUAAAAAUGCGUUU- ((((.(((..(((((((((...((((-.((((((((...)))..))))).))))...-----)))))))))..))).))))-.....(((.(......).)))............- ( -25.80, z-score = -2.58, R) >droWil1.scaffold_180708 721781 113 + 12563649 UUUAGUUGCGUUAGUUUAACGAGGAAUGAAGGGUCCGUUGUAUUUAAUUACUUUUUUGGUACAUUAGUUAAUAAAAGGAAACAAUUAAU--CAAUUAACAUUAUAAACUCGUUUU- .(((((((.(((((((((((..(((........)))..(((((..((.......))..)))))...))))......(....))))))))--))))))).................- ( -15.60, z-score = 0.45, R) >droEre2.scaffold_4929 1878584 94 - 26641161 UCCCAUUUUCUUAGCUUGGCAAUAAA-AUAUUAUUCAUCG-AACUAGUAAUUUAAUU-----CCAAGCCAACUAAAGGGGA-UAUUCACUGCAAAUGCAUUU-------------- ((((.(((..((.((((((...((((-.((((((((...)-)).))))).))))...-----)))))).))..))).))))-.......(((....)))...-------------- ( -19.90, z-score = -2.69, R) >droYak2.chr2L 1804987 96 - 22324452 UCCCAUUUUGUUAGCUUGGCAAUAAA-AUAUUGUUGAUUGAAACUAGUAAUUUACUU-----CCAAGCUAACUGAAGUGGAUAUUCUACAACAAAUGGGUUU-------------- .(((((((.((((((((((...((((-.(((((((......))).)))).))))...-----))))))))))....((((.....))))...)))))))...-------------- ( -30.20, z-score = -4.97, R) >droSec1.super_14 1778365 109 - 2068291 UCCCGUUUUCUUUGCUUGGCAAUAAA-AUAUUAUUCACAGAAACUAGUAACUUACUU-----CCAAGCUAACUGAAGGGGA-UAUUCAGUGCAACCACGGACUAAAAAUGCUUUUU ((((.(((..((.((((((...(((.-.((((((((...)))..)))))..)))...-----)))))).))..))).))))-.....(((.(......).)))............. ( -19.50, z-score = -0.59, R) >droSim1.chr2L 1818710 110 - 22036055 UCCCGUUUUCUUAGCUUGGCAAUAAA-AUAUUAUUCACAGAAACUAGUAAUUUACUU-----CCAAGCUAACUGAAGGGAAUUAUUCAGUGCAACAUCGGACUAAAAAUGCUUUUU ((((.(((..(((((((((...((((-.((((((((...)))..))))).))))...-----)))))))))..))))))).......(((.(......).)))............. ( -23.30, z-score = -2.25, R) >consensus UCCCGUUUUCUUAGCUUGGCAAUAAA_AUAUUAUUCACAGAAACUAGUAAUUUACUU_____CCAAGCUAACUGAAGGGGA_UAUUCAGUGCAACUACGGACUAAAAAUGCUUUU_ ((((.(((..(((((((((..........((((((..........))))))...........)))))))))..))).))))................................... (-11.66 = -11.87 + 0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:26 2011