
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,772,452 – 19,772,543 |
| Length | 91 |
| Max. P | 0.863262 |

| Location | 19,772,452 – 19,772,543 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 75.36 |
| Shannon entropy | 0.49764 |
| G+C content | 0.42173 |
| Mean single sequence MFE | -19.46 |
| Consensus MFE | -10.75 |
| Energy contribution | -10.51 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.863262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19772452 91 + 23011544 ---AAUCAUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUUGGGAAACUUUUGCCUUCCU-----AGCUCGACUCUUUCUACUCCGC ---..((..((--((((((((((((............))))))).))))))).(((((((.........)))))-----))...))............... ( -20.50, z-score = -1.69, R) >droSim1.chr2L 19429888 91 + 22036055 ---AAUCAUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUCGGGAAACUUUUGCCUUCCU-----AGUUCGACUCUUCCUGCUCCGC ---......((--((((((((((((............))))))).)))))))..((((((....(((.((....-----))..)))...))))))...... ( -20.10, z-score = -1.05, R) >droSec1.super_7 3377538 91 + 3727775 ---AAUCAUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUCGGGAAACUUUUGCCUUCCU-----AGCUCGACUCGUUCUGCUUAGC ---......((--((((((((((((............))))))).)))))))...(((((.........)))))-----(((..((....))..))).... ( -20.40, z-score = -0.61, R) >droYak2.chr2R 6235020 89 + 21139217 ---AAUCAUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUCGGGAAACUUUUGCCUUCCU-----AGCUCGACUC--UCUACUCCGC ---.....(((--((((((((((((............))))))).))))))(((.(((((.........)))))-----)))..))...--.......... ( -19.40, z-score = -1.13, R) >droEre2.scaffold_4845 18036486 91 - 22589142 ---AAUCAUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUCGGGAAAGUUUUGCCUUCCU-----AGCUCGACUCUUUCUACUCCGC ---......((--((((((((((((............))))))).)))))))...(((((((((..((......-----.))..)))).)))))....... ( -22.10, z-score = -1.72, R) >droAna3.scaffold_12916 7143149 96 - 16180835 ---AACCAUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUCCGGAAACUUUACUCUGUGUUGUGCGGCUCUGCUUUUUCUACUUCGC ---......((--((((((((((((............))))))).)))))))....(((((.......((((.....))))........)))))....... ( -19.56, z-score = -0.12, R) >dp4.chr4_group5 102012 91 + 2436548 ---AAUCAUCGCGUGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUUGGAAAACUUUUCUCCUAU-------GCUCCGCACUCCUCACACCCC ---.......(((.(((((((((((............))))))).))))((((..(((.........))).))-------))..))).............. ( -20.10, z-score = -2.11, R) >droPer1.super_5 4602727 91 + 6813705 ---AAUCAUCGCGUGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUUGGAAAACUUUUCUCCUAU-------GCUCCGCACUCCUCACACCCC ---.......(((.(((((((((((............))))))).))))((((..(((.........))).))-------))..))).............. ( -20.10, z-score = -2.11, R) >droVir3.scaffold_12963 1500120 76 + 20206255 ACACACACUCA--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUUGGCAAACUUUUUUUUUUUUUUAU----------------------- .....((..(.--((((((((((((............))))))).))))).)..))......................----------------------- ( -12.20, z-score = -1.03, R) >droMoj3.scaffold_6500 28397253 76 + 32352404 ACCCAGCCUCG--UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUUGGUAAACUUUUCUUUUUUUUUUU----------------------- .....(((.((--((((((((((((............))))))).)))))))...)))....................----------------------- ( -20.10, z-score = -3.79, R) >consensus ___AAUCAUCG__UGUGCACAUAUUUCGAUGUAAUUUAAUAUGUGGCACGCGUUCGGGAAACUUUUGCCUUCCU_____AGCUCGACUCUUUCUACUCCGC ........(((..((((((((((((............))))))).)))))....)))............................................ (-10.75 = -10.51 + -0.24)

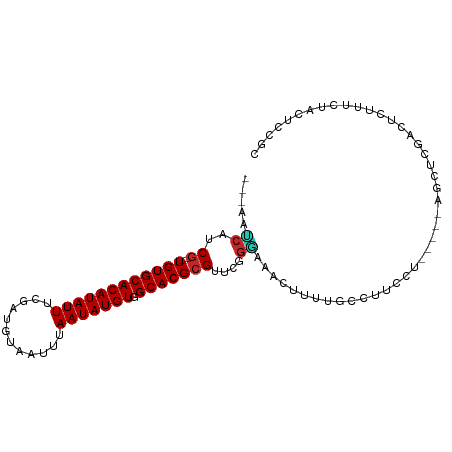
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:58 2011