
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,770,056 – 19,770,186 |
| Length | 130 |
| Max. P | 0.693488 |

| Location | 19,770,056 – 19,770,186 |
|---|---|
| Length | 130 |
| Sequences | 11 |
| Columns | 141 |
| Reading direction | forward |
| Mean pairwise identity | 87.07 |
| Shannon entropy | 0.27533 |
| G+C content | 0.38189 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.91 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.587812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
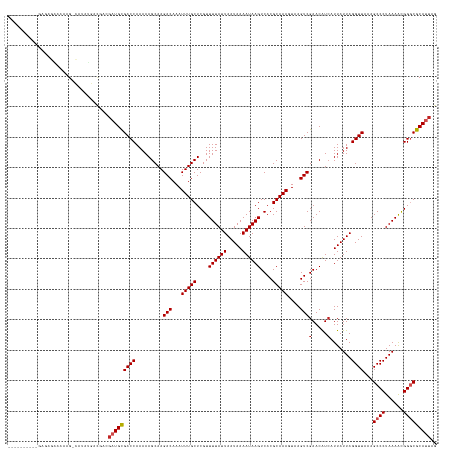
>dm3.chr2L 19770056 130 + 23011544 ----------GAGAGAAAAAG-CAUAUGACGACGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAACUAAGUCGAACGUCAUUAUUAGGUUAAAGCCCUAAAAUGGGCGCAUUAG ----------..........(-(..((((((....(((.........))).(((...(((((....((((((.....))))))....)))))....)))...))))))............((((......))))))..... ( -27.90, z-score = -2.02, R) >droSec1.super_7 3375148 130 + 3727775 ----------GAGAGAAAAAG-CAUAUGACGACGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAACUAAGUCGAACGUCAUUAUUAGGUUAAAGCCCUAAAACGGGCGCAUUAG ----------..........(-(..((((((....(((.........))).(((...(((((....((((((.....))))))....)))))....)))...))))))............((((......))))))..... ( -27.30, z-score = -1.81, R) >droSim1.chr2L 19426233 131 + 22036055 ----------GAGAGAAAAAGGCAUAUGACGACGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAACUAAGUCGAACGUCAUUAUUAGGUUAAAGCCCUAAAACGGGCGCAUUAG ----------...........((..((((((....(((.........))).(((...(((((....((((((.....))))))....)))))....)))...))))))............((((......))))))..... ( -27.10, z-score = -1.50, R) >droYak2.chr2R 6232764 130 + 21139217 ----------GAGAGAAAAAG-UAUAUCACGACGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAACUAAGUCGAACGUCAUUAUUAGGUUAAAGCCCUAAAACGGACGCAUUAG ----------...........-.......((((..((((.......((((((.....)))))).((((((....))))))......))))......))))..((((....(((((.......)))))....))))...... ( -22.10, z-score = -0.77, R) >droEre2.scaffold_4845 18033881 130 - 22589142 ----------GAGAGAAAAAG-CAUAUGACGACGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAACUAAGUCGAACGUCAUUAUUAGGUUAAAGCCCUAAAACGGACGCAUUAG ----------..........(-(......((((..((((.......((((((.....)))))).((((((....))))))......))))......))))...(((....(((((.......)))))....)))))..... ( -23.50, z-score = -1.05, R) >droAna3.scaffold_12916 7141378 128 - 16180835 -------AAGAGAUGGAGCAGCCUGACGA------UGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUAAUUAUUAGGUUAAAGCCCUAAAACGGGCGCAUUAG -------..........(((((((((..(------(((.........))..(((...(((((....((((((.....))))))....)))))....))).......))..)))))))...((((......))))))..... ( -29.00, z-score = -1.97, R) >dp4.chr4_group5 99610 124 + 2436548 ----------------GCGAG-UGAAUGUCGUGGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUCAUUAUUAGGUUAAAGCCCUAAAGCGGGCGCAUUAG ----------------((.((-(((..(((((....)))))..........(((...(((((....((((((.....))))))....)))))....))).....)))))...........((((......))))))..... ( -27.80, z-score = -1.03, R) >droPer1.super_5 4600339 124 + 6813705 ----------------GCGAG-UGAAUGUCGUGGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUCAUUAUUAGGUUAAAGCCCUAAAGCGGGCGCAUUAG ----------------((.((-(((..(((((....)))))..........(((...(((((....((((((.....))))))....)))))....))).....)))))...........((((......))))))..... ( -27.80, z-score = -1.03, R) >droWil1.scaffold_180708 8546591 121 - 12563649 -----------------CGAG---UAUGCUGAGGAUGUGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUCAUUAUUAGGUUAAAGCCCUAAAGCGGGCGCAUUAG -----------------(((.---(.((((...(.((((((.....((((((.....)))))).((((((....))))))..)))))))..))))).)))....................((((......))))....... ( -25.30, z-score = -0.95, R) >droMoj3.scaffold_6500 3950607 141 - 32352404 GUGUGUAGUUGUGUGUGUGUGCCUGAAGAGAUGGCUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUCAUUAUUAGGUUAAAGCCCUAAAGCGGGCGCAUUAG ................(((((((((...((..((((..((((.........(((...(((((....((((((.....))))))....)))))....)))(.....).......))))..))))))....)))))))))... ( -34.10, z-score = -0.66, R) >droGri2.scaffold_15126 7002529 130 + 8399593 ----------GUGUGAAAGUG-UAUCUGCCUUGGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUCAUUAUUAGGUUAAAGCCCUAAAGCGGGCGCAUUAG ----------((((......(-(((((.....))))))((((.........(((...(((((....((((((.....))))))....)))))....)))(.....).......))))...((((......))))))))... ( -26.90, z-score = -0.30, R) >consensus __________GAGAGAAAAAG_CAUAUGACGACGAUGCGACCAAAAAGCAAGACAAAUUUGCUCGUUUUAUUAAUAAAAUAAGUCACGCAAAGUAAGUCGAACAUCAUUAUUAGGUUAAAGCCCUAAAACGGGCGCAUUAG .................................(((((((((.........(((...(((((....((((((.....))))))....)))))....)))(.....).......))))...((((......))))))))).. (-22.45 = -22.91 + 0.46)


| Location | 19,770,056 – 19,770,186 |
|---|---|
| Length | 130 |
| Sequences | 11 |
| Columns | 141 |
| Reading direction | reverse |
| Mean pairwise identity | 87.07 |
| Shannon entropy | 0.27533 |
| G+C content | 0.38189 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.693488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
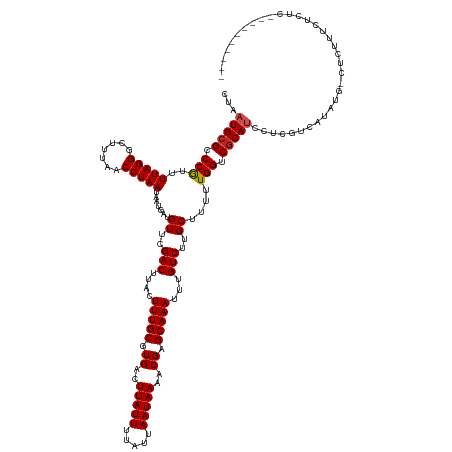
>dm3.chr2L 19770056 130 - 23011544 CUAAUGCGCCCAUUUUAGGGCUUUAACCUAAUAAUGACGUUCGACUUAGUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCGUCGUCAUAUG-CUUUUUCUCUC---------- .....((((((......))))............((((((..(((....(....)((((((((((.....)))))...((((((.......))))))....))))).))).))))))..)-)..........---------- ( -29.00, z-score = -2.13, R) >droSec1.super_7 3375148 130 - 3727775 CUAAUGCGCCCGUUUUAGGGCUUUAACCUAAUAAUGACGUUCGACUUAGUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCGUCGUCAUAUG-CUUUUUCUCUC---------- .....((((((......))))............((((((..(((....(....)((((((((((.....)))))...((((((.......))))))....))))).))).))))))..)-)..........---------- ( -29.70, z-score = -2.06, R) >droSim1.chr2L 19426233 131 - 22036055 CUAAUGCGCCCGUUUUAGGGCUUUAACCUAAUAAUGACGUUCGACUUAGUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCGUCGUCAUAUGCCUUUUUCUCUC---------- .....((((((......))))............((((((..(((....(....)((((((((((.....)))))...((((((.......))))))....))))).))).))))))..))...........---------- ( -29.30, z-score = -1.87, R) >droYak2.chr2R 6232764 130 - 21139217 CUAAUGCGUCCGUUUUAGGGCUUUAACCUAAUAAUGACGUUCGACUUAGUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCGUCGUGAUAUA-CUUUUUCUCUC---------- ...(((((.((((((((((.......)))))....)))((..(((..(((((((.((..(((((.....)))))..)).))))))).)))..)).....)).)))))............-...........---------- ( -27.40, z-score = -1.43, R) >droEre2.scaffold_4845 18033881 130 + 22589142 CUAAUGCGUCCGUUUUAGGGCUUUAACCUAAUAAUGACGUUCGACUUAGUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCGUCGUCAUAUG-CUUUUUCUCUC---------- ...(((((.((((((((((.......)))))....)))((..(((..(((((((.((..(((((.....)))))..)).))))))).)))..)).....)).)))))............-...........---------- ( -27.40, z-score = -1.49, R) >droAna3.scaffold_12916 7141378 128 + 16180835 CUAAUGCGCCCGUUUUAGGGCUUUAACCUAAUAAUUAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCA------UCGUCAGGCUGCUCCAUCUCUU------- .....((((((......))))...............................((.(((((((((.....)))))..(((.((((...(.....)..)))).)))..------..)))).)).))..........------- ( -24.60, z-score = -0.47, R) >dp4.chr4_group5 99610 124 - 2436548 CUAAUGCGCCCGCUUUAGGGCUUUAACCUAAUAAUGAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCCACGACAUUCA-CUCGC---------------- ..(((((((((......))))..............((((..(((((......(((.(((..((((..((........))..))))..))).))).....)))))))))...).))))..-.....---------------- ( -23.90, z-score = -0.71, R) >droPer1.super_5 4600339 124 - 6813705 CUAAUGCGCCCGCUUUAGGGCUUUAACCUAAUAAUGAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCCACGACAUUCA-CUCGC---------------- ..(((((((((......))))..............((((..(((((......(((.(((..((((..((........))..))))..))).))).....)))))))))...).))))..-.....---------------- ( -23.90, z-score = -0.71, R) >droWil1.scaffold_180708 8546591 121 + 12563649 CUAAUGCGCCCGCUUUAGGGCUUUAACCUAAUAAUGAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCACAUCCUCAGCAUA---CUCG----------------- ...((((((((......))))..............(((((..((((......(((.(((..((((..((........))..))))..))).))).....)))))))))....)))).---....----------------- ( -26.70, z-score = -2.38, R) >droMoj3.scaffold_6500 3950607 141 + 32352404 CUAAUGCGCCCGCUUUAGGGCUUUAACCUAAUAAUGAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAGCCAUCUCUUCAGGCACACACACACAACUACACAC ....(((((((......))))..............(((((((((((......(((.(((..((((..((........))..))))..))).))).....))))).)).)))).......)))................... ( -27.00, z-score = -0.89, R) >droGri2.scaffold_15126 7002529 130 - 8399593 CUAAUGCGCCCGCUUUAGGGCUUUAACCUAAUAAUGAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCCAAGGCAGAUA-CACUUUCACAC---------- ....(((((((......))))..............((((..(((((......(((.(((..((((..((........))..))))..))).))).....)))))))))....)))....-...........---------- ( -26.20, z-score = -0.86, R) >consensus CUAAUGCGCCCGUUUUAGGGCUUUAACCUAAUAAUGAUGUUCGACUUACUUUGCGUGACUUAUUUUAUUAAUAAAACGAGCAAAUUUGUCUUGCUUUUUGGUCGCAUCCUCGUCAUAUG_CUCUUUCUCUC__________ ...(((((.(((..(((((.......))))).......((..(((....(((((.((..(((((.....)))))..)).)))))...)))..))....))).))))).................................. (-22.06 = -22.25 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:57 2011