
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,769,813 – 19,769,905 |
| Length | 92 |
| Max. P | 0.529922 |

| Location | 19,769,813 – 19,769,905 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 54.37 |
| Shannon entropy | 0.94485 |
| G+C content | 0.61377 |
| Mean single sequence MFE | -32.69 |
| Consensus MFE | -10.24 |
| Energy contribution | -9.31 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 2.07 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.529922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
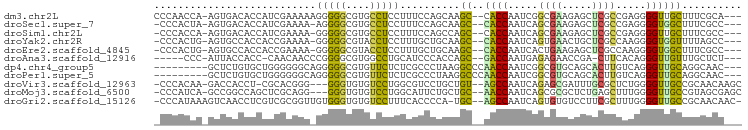
>dm3.chr2L 19769813 92 + 23011544 CCCAACCA-AGUGACACCAUCGAAAAAGGGGGCGUGCCUCCUUUCCAGCAAGC--CACCAAUCGGCGAAGAGCUCGCCGAGGGGUUGCUUUCGCA--- ........-.((((..........((((((((....)))))))).....((((--.(((..(((((((.....)))))))..))).)))))))).--- ( -36.70, z-score = -2.28, R) >droSec1.super_7 3374908 90 + 3727775 -CCCACUA-AGUGACACCAUCGAAAA-AGGGGCGUGCCUCCUUUCCAGCAAGC--CACCAAUCAGCGAAGAGCUCGCCGAGGGGUGGCUUUCGCC--- -.......-.((((..........((-(((((....)).))))).....((((--((((..((.((((.....)))).))..)))))))))))).--- ( -32.10, z-score = -1.50, R) >droSim1.chr2L 19425993 90 + 22036055 -CCCACCA-AGUGACACCAUCGAAAA-GGGGGCGUGCCUCCUUUCCAGCCAGC--CACCAAUCAGCGAAGAGCUCGCCGAGGGGUUGCUUUCGCC--- -.......-.((((.......(.(((-(((((....)))))))).)....(((--.(((..((.((((.....)))).))..))).))).)))).--- ( -29.80, z-score = -0.57, R) >droYak2.chr2R 6232541 90 + 21139217 -CCCACUG-AGUGCCACCACCGAAAA-GGGGGCGUACCUCCUUUGCUGCAAGC--CACCAAUCAGUGAACUGCUCGCCAAGGGGUGGUUUUAGCC--- -....(((-((.((((((.((..(((-(((((....))))))))((.(((...--(((......)))...)))..))...)))))))))))))..--- ( -31.40, z-score = -0.59, R) >droEre2.scaffold_4845 18033656 90 - 22589142 -CCCACUG-AGUGCCACCACCGAAAA-GGGGGCGUACCUCCUUUGCUGCAAGC--CACCAAUCACUGAAGAGCUCGCCAAGGGGUGGCUUUCGCC--- -......(-((.((((((.((..(((-(((((....))))))))...((.(((--(..((.....))..).))).))...)))))))).)))...--- ( -32.10, z-score = -0.75, R) >droAna3.scaffold_12916 7141189 85 - 16180835 -----CCC-AUUACCACC-CAACAACCCGGGGCGUGGCCUGCAUCCCACCAGC--GACCAAUGAGAGAACCGA-CUUCACAGGGUUGUUUGCUCU--- -----...-.........-.((((((((((.((((((........))))..))--..))..((.(((......-.))).))))))))))......--- ( -19.80, z-score = 0.68, R) >dp4.chr4_group5 99387 86 + 2436548 ---------GCUCUGUGCUGGGGGGCAGGGGGCGUGUUCUCUCGCCCUAAGGCCCAACCAAUCGGCGUGCAGCACUUGUCAGGGUUGCAGGCAAC--- ---------((.....))(((.((((..((((((.(....).))))))...))))..)))....((.((((((.((.....)))))))).))...--- ( -38.40, z-score = -1.17, R) >droPer1.super_5 4600116 86 + 6813705 ---------GCUCUGUGCUGGGGGGCAGGGGGCGUGUUCUCUCGCCCUAAGGCCCAACCAAUCGGCGUGCAGCACUUGUCAGGGUUGCAGGCAAC--- ---------((.....))(((.((((..((((((.(....).))))))...))))..)))....((.((((((.((.....)))))))).))...--- ( -38.40, z-score = -1.17, R) >droVir3.scaffold_12963 18703134 90 - 20206255 -CCCACAA-GACCACCU-CGCACGGG---GGGUGUGUCCUGGCGUCCUGCUGU--AGCCAAUCAGAGCGAUUUGCGCUCUGGGGUUGCCGCAACAAGC -....((.-((((((((-(......)---))))).))).))((....(((.((--((((...(((((((.....))))))).)))))).)))....)) ( -39.40, z-score = -1.72, R) >droMoj3.scaffold_6500 3950397 91 - 32352404 -CCCAUCA-GCCGGCCAGCUCGCAGG---GGGUGUGUCCUGGCAUUCUGCUGC--AACCAAUCAGCGCGCUCUGAGCUUUGGGGUUGCCGUAGCGAGC -.......-(((((((((..((((..---...))))..)))))...((((.((--((((...(((.((.......)).))).)))))).)))))).)) ( -32.70, z-score = 1.38, R) >droGri2.scaffold_15126 7002248 93 + 8399593 -CCCAUAAAGUCAACCUCGUCGCGGUUGUGGGUGUGUCCUUUCACCCCA-UGC--AGCCAAUCAGUGUGUCCUUCGCUUUGGGGUUGCCGCAACAAC- -.................((.(((...(((((.(((......)))))))-)((--((((...(((.(((.....))).))).))))))))).))...- ( -28.80, z-score = -1.37, R) >consensus _CCCACCA_AGUGACACCAUCGAAAA_GGGGGCGUGCCCUCUCUCCCGCAAGC__AACCAAUCAGCGAACAGCUCGCCUAGGGGUUGCUUUCGCC___ ...........................(((((....)))))..........((..((((.....((((.....)))).....)))))).......... (-10.24 = -9.31 + -0.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:55 2011