
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,769,036 – 19,769,129 |
| Length | 93 |
| Max. P | 0.758789 |

| Location | 19,769,036 – 19,769,129 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 85.89 |
| Shannon entropy | 0.29335 |
| G+C content | 0.52181 |
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -26.01 |
| Energy contribution | -25.76 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
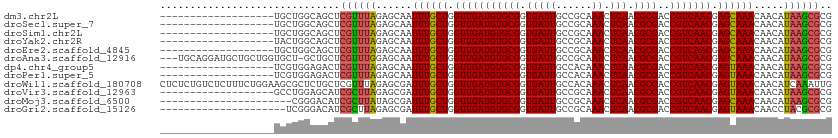
>dm3.chr2L 19769036 93 - 23011544 -------------------UGCUGGCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG -------------------.(((....((((.....))))..((((((.(((((((((((((....))))))...((.....)).))))))).)))))).......)))... ( -34.60, z-score = -1.86, R) >droSec1.super_7 3374144 93 - 3727775 -------------------UGCUGGCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG -------------------.(((....((((.....))))..((((((.(((((((((((((....))))))...((.....)).))))))).)))))).......)))... ( -34.60, z-score = -1.86, R) >droSim1.chr2L 19425214 93 - 22036055 -------------------UGCUGGCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG -------------------.(((....((((.....))))..((((((.(((((((((((((....))))))...((.....)).))))))).)))))).......)))... ( -34.60, z-score = -1.86, R) >droYak2.chr2R 6231749 93 - 21139217 -------------------UACUGGCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG -------------------.....((.((((.....))))..((((((.(((((((((((((....))))))...((.....)).))))))).))))))..........)). ( -34.00, z-score = -2.25, R) >droEre2.scaffold_4845 18032865 93 + 22589142 -------------------UGCUGGCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG -------------------.(((....((((.....))))..((((((.(((((((((((((....))))))...((.....)).))))))).)))))).......)))... ( -34.60, z-score = -1.86, R) >droAna3.scaffold_12916 7140220 108 + 16180835 ---UGCAGGAUGCUGCUGGUGCU-GCUGCUCGUUUGGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG ---.((((....))))..(((((-..(((((.....))))).((((((.(((((((((((((....))))))...((.....)).))))))).)))))).......))))). ( -43.10, z-score = -1.80, R) >dp4.chr4_group5 98206 93 - 2436548 -------------------UCGUGGAGACUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCACAAACUCAACGCGACCGUCAACGAGUAAACAACAUAAGCGCG -------------------.((((..(.(((.....))))..((((((.(((((((((((.(((((......)).))).))))..))))))).)))))).........)))) ( -26.50, z-score = -0.57, R) >droPer1.super_5 4598948 93 - 6813705 -------------------UCGUGGAGACUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCACAAACUCAACGCGACCGUCAACGAGUAAACAACAUAAGCGCG -------------------.((((..(.(((.....))))..((((((.(((((((((((.(((((......)).))).))))..))))))).)))))).........)))) ( -26.50, z-score = -0.57, R) >droWil1.scaffold_180708 8545416 112 + 12563649 CUCUCUGUCUCUUUCUGGAAGCGCUCUGCUCGUUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCACAAACUCAACGCGACCGUCAACGAGUAAACAACAUCAAAUUG ......(..(((....)))..)((((((......))))))(.((((((.(((((((((((.(((((......)).))).))))..))))))).)))))))............ ( -31.10, z-score = -1.10, R) >droVir3.scaffold_12963 18702128 93 + 20206255 -------------------GCCUGGAGCAUCGCUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGUAAACAACAUAAGCGCG -------------------((.....))..((((((......((((((.(((((((((((((....))))))...((.....)).))))))).)))))).....)))))).. ( -30.50, z-score = -0.89, R) >droMoj3.scaffold_6500 3948653 90 + 32352404 ----------------------CGGGACAUCGCUUAUAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG ----------------------........(((((((...(.((((((.(((((((((((((....))))))...((.....)).))))))).)))))))...))))))).. ( -32.20, z-score = -2.11, R) >droGri2.scaffold_15126 7001180 91 - 8399593 ---------------------UCGGGACAUCGCUUAGAGCGAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGUAAACAACCUACGCGCG ---------------------..(((...((((.....))))((((((.(((((((((((((....))))))...((.....)).))))))).))))))...)))....... ( -30.70, z-score = -1.22, R) >consensus ___________________UGCUGGCAGCUCGUUUAGAGCAAUUUGCUGGUUGAUGUGCGGUGAUUGCCGCAAACUCAACGCGACCGUCAACGAGCAAACAACAUAAGCGCG ..............................((((((......((((((.(((((((((((.(((((......)).))).))))..))))))).)))))).....)))))).. (-26.01 = -25.76 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:54 2011