| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,756,919 – 19,757,009 |
| Length | 90 |
| Max. P | 0.577584 |

| Location | 19,756,919 – 19,757,009 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 52.88 |
| Shannon entropy | 0.91110 |
| G+C content | 0.46764 |
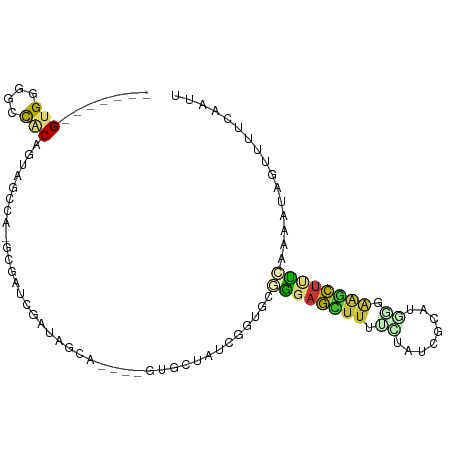
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -5.46 |
| Energy contribution | -5.10 |
| Covariance contribution | -0.36 |
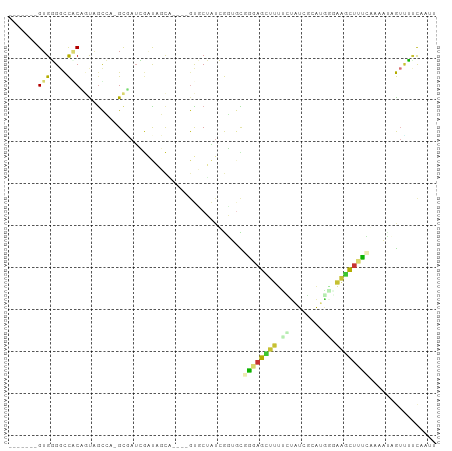
| Combinations/Pair | 2.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
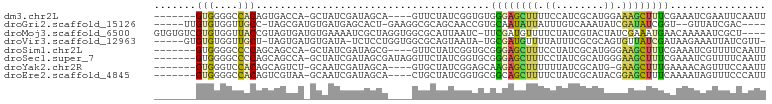
>dm3.chr2L 19756919 90 + 23011544 -------GUGGGGCCACAGUGACCA-GCUAUCGAUAGCA----GUUCUAUCGGUGUGGGAGCUUUUCCAUCGCAUGGAAAGCUUUCGAAAUCGAAUUCAAUU -------(((....)))(((..(((-..(((((((((..----...))))))))))))..)))(((((((...)))))))(..((((....))))..).... ( -26.70, z-score = -1.30, R) >droGri2.scaffold_15126 6989213 89 + 8399593 -----UUGUGUGGUUGCC-UAGCGAUGUGAUGAGCACU-GAAGGCGCAGCAACCGUGCAAUAUUAUUUGUCAAAUAUCGAUAUCGGU--GUUAUCGAC---- -----(((..(((((((.-..(((..(((.....))).-.....))).)))))))..)))........(((.(((((((....))))--)))...)))---- ( -26.80, z-score = -1.31, R) >droMoj3.scaffold_6500 3934081 97 - 32352404 GUGUGUCGUGUGGUUACCGUAGUGAUGUGAAAAUCGCUAGGUGGCGCAUUAAUC-UUCGAUGUUUUCUAUCGUACUAUCGAAAUGAACAAAAAUCGCU---- .(((.(((((((((((((.(((((((......))))))))))))).))).....-((((((((..........)).))))))))))))).........---- ( -24.80, z-score = -1.60, R) >droVir3.scaffold_12963 18688845 93 - 20206255 -----GUGUGUGGUUGCU-UAGUGAUGUGAUA-UCUCCUGGUGGCGCAGUAAUA-UGCGAUGUUUUAUUUCGCGCAGUGUUAUCGAUAAGAAAUUAUCGUU- -----.((((((((..((-..(.((((...))-)).)..))..))(((......-)))............)))))).......((((((....))))))..- ( -24.40, z-score = -1.09, R) >droSim1.chr2L 19413018 90 + 22036055 -------GUGGGGCCCCAGCAGCCA-GCUAUCGAUAGCG----GUUCUAUCGGUGCGGGAGCUUUCCUAUCGCAUGGGAAGCUUUCGAAAUCGUUUUCAAUU -------.(((.((....))..)))-((.((((((((..----...))))))))))(..((((((((........))))))))..)((((....)))).... ( -32.80, z-score = -1.74, R) >droSec1.super_7 3362384 94 + 3727775 -------GUGGGGCCCCAGCAGCCA-GCUAUCGAUAGCGAUAGGUUCUAUCGGUGCGGGAGCUUUCCUAUCGCAUGGGAAGCUUUCGAAAUCGUUUUCAAUU -------.(((....)))(((.((.-((((....))))(((((...))))))))))(..((((((((........))))))))..)((((....)))).... ( -34.50, z-score = -1.94, R) >droYak2.chr2R 6219101 89 + 21139217 -------GUGGGUCCACAGCAGUCU-GCAAUCGAUAGCA----GUGCUAUCGGAGCAAGAGCUUUUUUAUCGCAUG-GAAGCUUUGAAAACAGUUUCCAAUU -------((((......(((..(((-((..((((((((.----..)))))))).)).))))))......)))).((-((((((.(....).))))))))... ( -27.90, z-score = -2.31, R) >droEre2.scaffold_4845 18021088 90 - 22589142 -------GUGGGGCCACAGUCGUAA-GCAAUCGAUAGCA----CUGCUAUCGGUGCGGCAGCUUUUCUAUCGCAUACGGAGCUUUCAAAAUAGUUUCCCAUU -------((((((((((....))..-(((.((((((((.----..))))))))))))))((((((............)))))).............))))). ( -27.10, z-score = -1.92, R) >consensus _______GUGGGGCCACAGUAGCCA_GCGAUCGAUAGCA____GUGCUAUCGGUGCGGGAGCUUUUCUAUCGCAUGGGAAGCUUUCAAAAUAGUUUUCAAUU .......(((....))).......................................((((((((.((........)).))))))))................ ( -5.46 = -5.10 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:51 2011