| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,702,556 – 19,702,640 |
| Length | 84 |
| Max. P | 0.556954 |

| Location | 19,702,556 – 19,702,640 |
|---|---|
| Length | 84 |
| Sequences | 7 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 73.75 |
| Shannon entropy | 0.48431 |
| G+C content | 0.54385 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -14.17 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19702556 84 - 23011544 CCCCUCCCCCCCCUUAC-------GCGCAUACGUCAGGAUAUCGCCAGGACGCAUGGUUCUGGAUCGCAGCUGCGUAUGCAACGAUUUUUU .................-------..(((((((.(((.......(((((((.....))))))).......))))))))))........... ( -25.04, z-score = -1.49, R) >droSim1.chr2L 19370529 75 - 22036055 ---------CCCCUCAC-------GCGCAUACGUCAGGAUAUCGCCAGGACGCGAGGUUCUGGAUCGCAGCUGCGUAUGCAACGAAUUUUU ---------........-------..(((((((.(((.......(((((((.....))))))).......))))))))))........... ( -25.04, z-score = -1.42, R) >droSec1.super_7 3317508 74 - 3727775 ---------CCCCUCAC-------GCGCAUACGUCAGGAUAUCGCCAGGACGCGAGGUUCUGGAUCGCAGCUGCGUAUGCAACGAUUUUU- ---------........-------..(((((((.(((.......(((((((.....))))))).......))))))))))..........- ( -25.04, z-score = -1.38, R) >droYak2.chr2R 6175373 75 - 21139217 ---------CCCCUCAC-------GCGCAUACGUCAGGAUAUCGUCAGGACGCGUGGUUCUGUAUCGCAGCUGCGUAUGCAACGAUUUUUU ---------........-------..(((((((.(((.....((.((((((.....))))))...))...))))))))))........... ( -21.30, z-score = -0.48, R) >droEre2.scaffold_4845 17979076 75 + 22589142 ---------CCCCUCAC-------GCGCAUACGUCAGGAUAUCGCCAGGACGCGUGGUUCUGCAUCGCAGCUGCGUAUGCAACGAUUUUUU ---------........-------..(((((((.(((......((((.(...).)))).((((...))))))))))))))........... ( -20.70, z-score = 0.09, R) >droAna3.scaffold_12916 7086154 78 + 16180835 -----UCCCUCACACAC-------ACACAUACGUCAGGAUAUGGCCUUAGCUUGUGUUGGUAAGACGCAGCUGCGUAUGCAACGAUUUUU- -----............-------...(((((((.(((......))).((((.(((((.....))))))))))))))))...........- ( -17.80, z-score = -0.15, R) >dp4.chr4_group5 1796168 87 - 2436548 -CCCCUCCCUGGCACACUGCCACGUCACAUACAUCAGAGAACAUCAGAGACGAGACUUUAC---CCGCAGCUGCGUAUGCAACGAUUUCCA -........((((.....))))(((..(((((...((((....((......))..))))..---..((....)))))))..)))....... ( -16.00, z-score = -0.73, R) >consensus _________CCCCUCAC_______GCGCAUACGUCAGGAUAUCGCCAGGACGCGUGGUUCUGGAUCGCAGCUGCGUAUGCAACGAUUUUUU ..........................(((((((.(((........((((((.....))))))........))))))))))........... (-14.17 = -15.08 + 0.90)

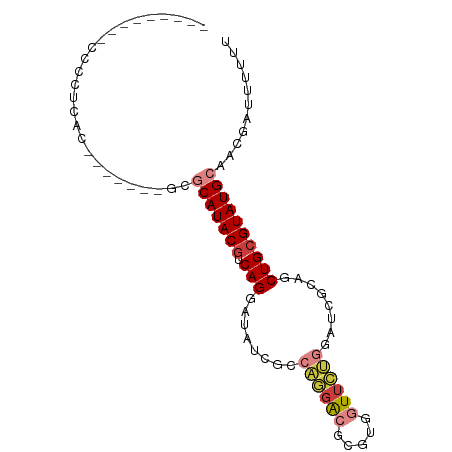
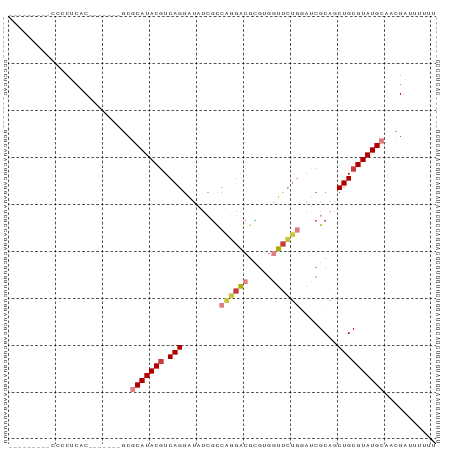
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:48 2011