| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,698,958 – 19,699,051 |
| Length | 93 |
| Max. P | 0.811764 |

| Location | 19,698,958 – 19,699,051 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 88.30 |
| Shannon entropy | 0.18952 |
| G+C content | 0.47161 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -16.60 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
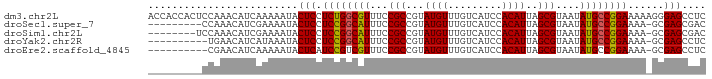
>dm3.chr2L 19698958 93 + 23011544 ACCACCACUCCAAACAUCAAAAAUACUCCUCUGGCGUUUCCGCCGUAUGUUUGUCAUCCACAUUAGCGUAAUAUGCCGGAAAAAGGGAGCCUC .........................((((((((((((...(((...((((.........))))..)))....)))))))).....)))).... ( -19.50, z-score = -1.37, R) >droSec1.super_7 3313893 83 + 3727775 ---------CCAAACAUCGAAAAUACUCCUCCGGCAUUUCCGCCGUAUGUUUGUCAUCCACAUUAGCGUAAUAUGCCGGAAAA-GCGAGCGAC ---------.......(((......((..((((((((...(((...((((.........))))..)))....))))))))..)-)....))). ( -20.50, z-score = -2.02, R) >droSim1.chr2L 19366942 84 + 22036055 --------UCCAAACAUCGAAAAUACUCCUCCGGCAUUUCCGCCGUAUGUUUGUCAUCCACAUUAGCGUAAUAUGCCGGAAAA-GCGAGCGAC --------........(((......((..((((((((...(((...((((.........))))..)))....))))))))..)-)....))). ( -20.50, z-score = -2.00, R) >droYak2.chr2R 6171786 82 + 21139217 ----------UGAACAUCAUAAAUACUCCUCCGGCAUUUCCGCCGUAUGUUUGUCAUCCACAUUAGCGUAAUAUGCCGGAAAA-GCGAGCCUC ----------...............((((((((((((...(((...((((.........))))..)))....))))))))...-).))).... ( -19.10, z-score = -1.91, R) >droEre2.scaffold_4845 17975501 82 - 22589142 ----------CGAACAUCAAAAAUACUCAUCCGUCGUUUCCGCCGUAUGUUUGUCAUCCACAUUAGCGUAAUAUGCCGGAAAA-GCGAGCCUC ----------.....................(((..((((((.(((((...((..(.......)..))..))))).)))))).-)))...... ( -13.60, z-score = -0.30, R) >consensus _________CCAAACAUCAAAAAUACUCCUCCGGCAUUUCCGCCGUAUGUUUGUCAUCCACAUUAGCGUAAUAUGCCGGAAAA_GCGAGCCUC .............................((((((((...(((...((((.........))))..)))....))))))))............. (-16.60 = -16.40 + -0.20)
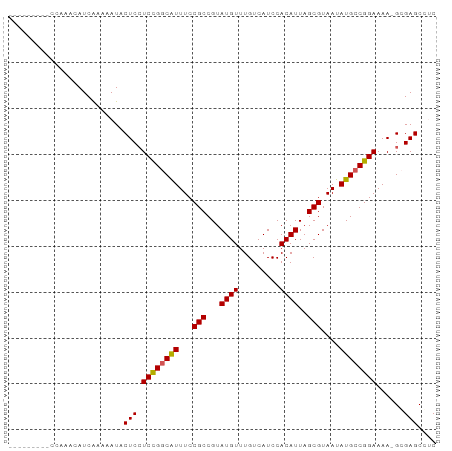
| Location | 19,698,958 – 19,699,051 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 88.30 |
| Shannon entropy | 0.18952 |
| G+C content | 0.47161 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -19.08 |
| Energy contribution | -20.08 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
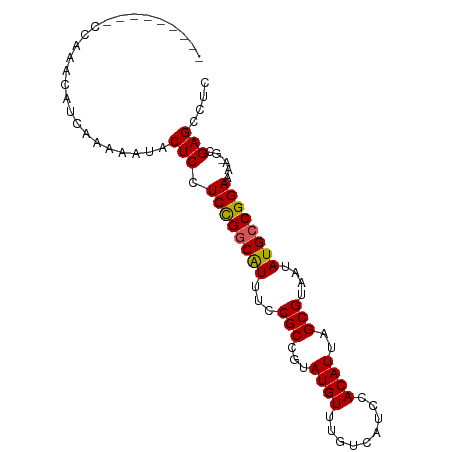
>dm3.chr2L 19698958 93 - 23011544 GAGGCUCCCUUUUUCCGGCAUAUUACGCUAAUGUGGAUGACAAACAUACGGCGGAAACGCCAGAGGAGUAUUUUUGAUGUUUGGAGUGGUGGU ...((..((...(((((((((....(((....))).....((((.((((((((....))))......)))).))))))).)))))).))..)) ( -26.40, z-score = -1.05, R) >droSec1.super_7 3313893 83 - 3727775 GUCGCUCGC-UUUUCCGGCAUAUUACGCUAAUGUGGAUGACAAACAUACGGCGGAAAUGCCGGAGGAGUAUUUUCGAUGUUUGG--------- ((((...((-(((((((((((....((((.((((.........))))..))))...))))))))))))).....))))......--------- ( -29.70, z-score = -3.20, R) >droSim1.chr2L 19366942 84 - 22036055 GUCGCUCGC-UUUUCCGGCAUAUUACGCUAAUGUGGAUGACAAACAUACGGCGGAAAUGCCGGAGGAGUAUUUUCGAUGUUUGGA-------- ((((...((-(((((((((((....((((.((((.........))))..))))...))))))))))))).....)))).......-------- ( -29.70, z-score = -2.94, R) >droYak2.chr2R 6171786 82 - 21139217 GAGGCUCGC-UUUUCCGGCAUAUUACGCUAAUGUGGAUGACAAACAUACGGCGGAAAUGCCGGAGGAGUAUUUAUGAUGUUCA---------- .......((-(((((((((((....((((.((((.........))))..))))...)))))))))))))..............---------- ( -27.80, z-score = -3.06, R) >droEre2.scaffold_4845 17975501 82 + 22589142 GAGGCUCGC-UUUUCCGGCAUAUUACGCUAAUGUGGAUGACAAACAUACGGCGGAAACGACGGAUGAGUAUUUUUGAUGUUCG---------- .......((-((.((((.(((((((...)))))))................((....)).)))).))))..............---------- ( -16.60, z-score = 0.36, R) >consensus GAGGCUCGC_UUUUCCGGCAUAUUACGCUAAUGUGGAUGACAAACAUACGGCGGAAAUGCCGGAGGAGUAUUUUUGAUGUUUGG_________ ...((((.....(((((((((....((((.((((.........))))..))))...)))))))))))))........................ (-19.08 = -20.08 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:48 2011