| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,827,031 – 1,827,149 |
| Length | 118 |
| Max. P | 0.950137 |

| Location | 1,827,031 – 1,827,149 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.70 |
| Shannon entropy | 0.53090 |
| G+C content | 0.42966 |
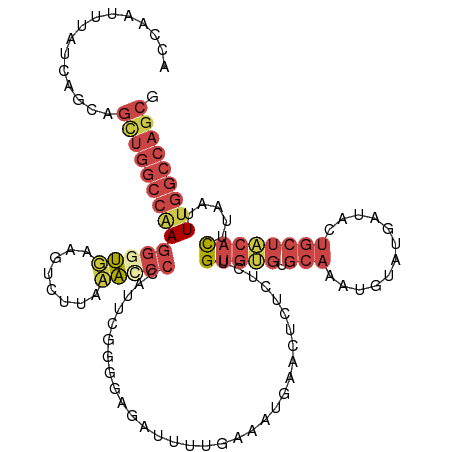
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -17.55 |
| Energy contribution | -19.55 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
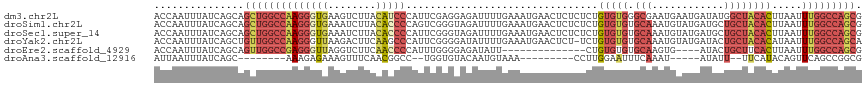
>dm3.chr2L 1827031 118 + 23011544 ACCAAUUUAUCAGCAGCUGGCCAAGGGUGAAGUCUUACAUCCCAUUCGAGGAGAUUUUGAAAUGAACUCUCUCUGUGUGGGCGAAUGAAUGAUAUGGCUACACUUAAUUUGGCCAGCG ...............((((((((((((((.((((.((((((((((.((..((((.(((.....))).))))..)).))))).......))).)).)))).))))...)))))))))). ( -40.71, z-score = -3.20, R) >droSim1.chr2L 1814012 118 + 22036055 ACCAAUUUAUCAGCAGCUGGCCAAGGGUGAAAUCUUACACCCCAGUCGGGUAGAUUUUGAAAUGAACUCUCUCUGUGUGUGCAAAUGUAUGAUGCUGCUACACUUAAUUUGGCCAGCG ...............((((((((((((((........))))).....(((.(((.(((.....))).)))))).(((((.(((.((.....))..)))))))).....))))))))). ( -35.80, z-score = -1.89, R) >droSec1.super_14 1773741 118 + 2068291 ACCAAUUUAUCAGCAGCUGGCCAAGGGUGAAAUCUUACACCCCAUUCGGGUAGAUUUUGAAAUGAACUCUCUCUGUGUGUGCAAAUGUAUGAUGCUGCUACACUUAAUUUGGCCAGCG ...............((((((((((((((........))))).....(((.(((.(((.....))).)))))).(((((.(((.((.....))..)))))))).....))))))))). ( -35.80, z-score = -2.22, R) >droYak2.chr2L 1800273 117 + 22324452 ACCAAUUUAUCAGCUGUUGGCCAAGGGUUAAGACUUCAAGCCCAUUCGGGGAUAUUUUGAAAUGAACUCU-UCUGUGUGUGCAAAUGUAUGAUACUGCUACACAUAAUUUGGCCAGCA ...............((((((((((((((.....(((((((((.....)))....))))))...))))).-..((((((((((............))).)))))))..))))))))). ( -34.50, z-score = -1.90, R) >droEre2.scaffold_4929 1872058 100 + 26641161 ACCAAUUUAUCAGCAGUUGGCCGAGGGUUAGGUCUUCAACCCCAUUUGGGGAGAUAUU--------------CUGUGUGUGCAAGUG----AUACUGCUUCACUUAAUUUGGCCAGCG ...............((((((((((((....((((....((((....))))))))...--------------))........(((((----(.......))))))..)))))))))). ( -30.80, z-score = -1.32, R) >droAna3.scaffold_12916 417597 92 - 16180835 AUUAAUUUAUCAGC--------AAAGAGAAAGUUUCAACGGCC--UGGUGUACAAUGUAAA---------CCUUGGAAUUUCAAAU-----AUAUU--UUCAUACAGUUCAGCCGGCG ............((--------...((((...))))..(((((--(((((...((((((..---------..((((....)))).)-----)))))--..))).)))....)))))). ( -13.00, z-score = 0.48, R) >consensus ACCAAUUUAUCAGCAGCUGGCCAAGGGUGAAGUCUUAAACCCCAUUCGGGGAGAUUUUGAAAUGAACUCUCUCUGUGUGUGCAAAUGUAUGAUACUGCUACACUUAAUUUGGCCAGCG ...............((((((((((((((........)))))................................(((((.(((............)))))))).....))))))))). (-17.55 = -19.55 + 2.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:24 2011