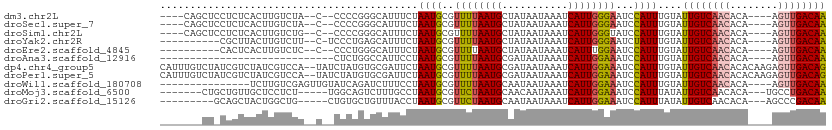
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,684,135 – 19,684,234 |
| Length | 99 |
| Max. P | 0.883290 |

| Location | 19,684,135 – 19,684,234 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.54107 |
| G+C content | 0.36782 |
| Mean single sequence MFE | -20.99 |
| Consensus MFE | -11.82 |
| Energy contribution | -11.96 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
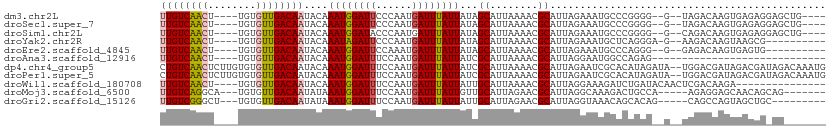
>dm3.chr2L 19684135 99 + 23011544 UUGUCAACU----UGUGUUGACAAUACAAAUGGAUUCCCAAUGAUUUAUUAUAGCAUUAAAACGCAUUAGAAAUGCCCGGGG--G--UAGACAAGUGAGAGGAGCUG---- ((((((((.----...))))))))..((..((.((((((((((...........)))).....((((.....))))..))))--)--)...))..))..........---- ( -21.50, z-score = -0.73, R) >droSec1.super_7 3299086 99 + 3727775 UUGUCAACU----UGUGUUGACAAUACAAAUGGAUUCCCAAUGAUUUAUUAUAGCAUUAAAACGCAUUAGAAAUGCCCGGGG--G--UAGACAAGUGAGAGGAGCUG---- ((((((((.----...))))))))..((..((.((((((((((...........)))).....((((.....))))..))))--)--)...))..))..........---- ( -21.50, z-score = -0.73, R) >droSim1.chr2L 19352195 99 + 22036055 UUGUCAACU----UGUGUUGACAAUACAAAUGGAUACCCAAUGAUUUAUUAUAGCAUUAAAACGCAUUAGAAAUGCCCGGGG--G--CAGACAAGUGAGAGGAGCUG---- ((((((((.----...))))))))......(((....)))...........((((.((......((((.(...((((....)--)--))..).))))....))))))---- ( -22.30, z-score = -1.07, R) >droYak2.chr2R 6157709 94 + 21139217 UUGUCAACU----UGUGUUGACAAUACAAAUAGAUUCCCAAUGAUUUAUUAUAGCAUUAAAACGCAUUAGAAAUGCUCAGGGA-G--AAGACAAGUAAGCG---------- ((((((((.----...))))))))..........(((((((((...........)))).....((((.....))))...))))-)--..............---------- ( -18.90, z-score = -1.86, R) >droEre2.scaffold_4845 17960732 93 - 22589142 UUGUCAACU----UGUGUUGACAAUACAAAUGGAUUCCAAAUGAUUUAUUAUAGCAUUAAAACGCAUUAGAAAUGCCCAGGG--G--GAGACAAGUGAGUG---------- ((((((((.----...))))))))..((..((..((((...((.....(((......)))...((((.....)))).))..)--)--))..))..))....---------- ( -19.80, z-score = -1.03, R) >droAna3.scaffold_12916 7068511 78 - 16180835 UUGUCAACU----UGUGUUGACAAUACAAAUGGAUUUCCAAUGAUUUAUUAUCGCAUUAAAACGCAUUAGGAAUGGCCAGAG----------------------------- ((((((((.----...))))))))......(((..((((...(((.....)))((........))....))))...)))...----------------------------- ( -19.70, z-score = -2.76, R) >dp4.chr4_group5 1771220 109 + 2436548 CUGUCAACUCUUGUGUGUUGACAAUACAAAUGGAUUUCCAAUGAUUUAUUAUCGCAUUAAAACGCAUUAGAAUCGCACAUAGAUA--UGGACGAUAGACGAUAGACAAAUG .(((((((.(....).))))))).......(((....))).((.(((((..((((........))......((((..(((....)--))..)))).))..))))))).... ( -23.50, z-score = -1.55, R) >droPer1.super_5 6180740 109 + 6813705 CUGUCAACUCUUGUGUGUUGACAAUACAAAUGGAUUUCCAAUGAUUUAUUAUCGCAUUAAAACGCAUUAGAAUCGCACAUAGAUA--UGGACGAUAGACGAUAGACAAAUG .(((((((.(....).))))))).......(((....))).((.(((((..((((........))......((((..(((....)--))..)))).))..))))))).... ( -23.50, z-score = -1.55, R) >droWil1.scaffold_180708 8431839 92 - 12563649 UUGUCAACU----UGUGUUGACAAUACAAAUGGAUUUCCAAUGAUUUAUUAUUGCAUUAAAACGCAUUAGGAAAGAUCUGAUACAACUCGACAAGA--------------- (((((...(----((((((...........(((....)))..(((((.(((.(((........))).)))...))))).)))))))...)))))..--------------- ( -18.10, z-score = -1.57, R) >droMoj3.scaffold_6500 3863381 96 - 32352404 UUGUCAGGCA---UGUGUUGACAAUAUAAAUGGAUUUCCAAUGAUUUAUUGUUGCAUUAGAACGCAUUAGGCAAAGACUGCCA-----AGAGGAGCAACAGCAG------- .(((...(((---(((((((((((((....(((....)))......))))))).......)))))))..((((.....)))).-----......))....))).------- ( -20.21, z-score = -0.17, R) >droGri2.scaffold_15126 6921422 94 + 8399593 UUGUCGGGCU---UGUGUUGACAAUAUAAAUGGAUUUCCAAUGAUUUAUUAUUGCAUUAGAACGCAUUAGGUAAACAGCACAG-----CAGCCAGUAGCUGC--------- ..........---(((((((..........(((....)))....((((((..(((........)))...)))))))))))))(-----((((.....)))))--------- ( -21.90, z-score = -0.46, R) >consensus UUGUCAACU____UGUGUUGACAAUACAAAUGGAUUUCCAAUGAUUUAUUAUAGCAUUAAAACGCAUUAGAAAUGCCCAGGG__G__UAGACAAGUGAGAG_AG_______ ((((((((........))))))))....((((((((......))))))))...((........)).............................................. (-11.82 = -11.96 + 0.14)
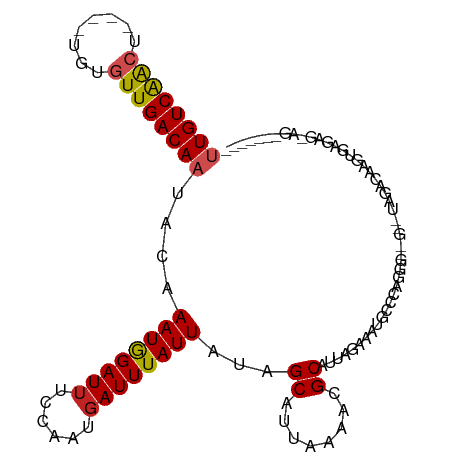
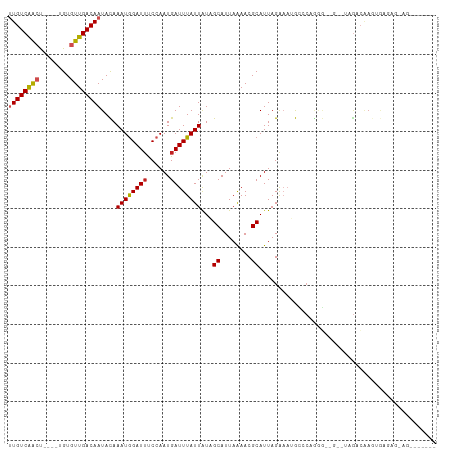
| Location | 19,684,135 – 19,684,234 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.89 |
| Shannon entropy | 0.54107 |
| G+C content | 0.36782 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -10.88 |
| Energy contribution | -10.89 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
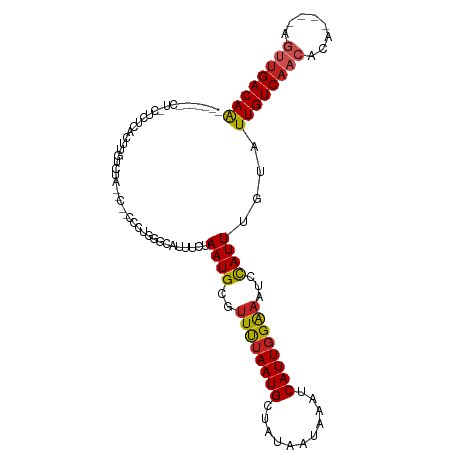
>dm3.chr2L 19684135 99 - 23011544 ----CAGCUCCUCUCACUUGUCUA--C--CCCCGGGCAUUUCUAAUGCGUUUUAAUGCUAUAAUAAAUCAUUGGGAAUCCAUUUGUAUUGUCAACACA----AGUUGACAA ----..................((--(--.....((.((((((((((.((((............))))))))))))))))....)))((((((((...----.)))))))) ( -19.30, z-score = -1.18, R) >droSec1.super_7 3299086 99 - 3727775 ----CAGCUCCUCUCACUUGUCUA--C--CCCCGGGCAUUUCUAAUGCGUUUUAAUGCUAUAAUAAAUCAUUGGGAAUCCAUUUGUAUUGUCAACACA----AGUUGACAA ----..................((--(--.....((.((((((((((.((((............))))))))))))))))....)))((((((((...----.)))))))) ( -19.30, z-score = -1.18, R) >droSim1.chr2L 19352195 99 - 22036055 ----CAGCUCCUCUCACUUGUCUG--C--CCCCGGGCAUUUCUAAUGCGUUUUAAUGCUAUAAUAAAUCAUUGGGUAUCCAUUUGUAUUGUCAACACA----AGUUGACAA ----..........((..((..((--(--((....((((.....))))((......))..............)))))..))..))..((((((((...----.)))))))) ( -20.10, z-score = -1.11, R) >droYak2.chr2R 6157709 94 - 21139217 ----------CGCUUACUUGUCUU--C-UCCCUGAGCAUUUCUAAUGCGUUUUAAUGCUAUAAUAAAUCAUUGGGAAUCUAUUUGUAUUGUCAACACA----AGUUGACAA ----------.(((((........--.-....)))))((((((((((.((((............)))))))))))))).........((((((((...----.)))))))) ( -18.62, z-score = -1.96, R) >droEre2.scaffold_4845 17960732 93 + 22589142 ----------CACUCACUUGUCUC--C--CCCUGGGCAUUUCUAAUGCGUUUUAAUGCUAUAAUAAAUCAUUUGGAAUCCAUUUGUAUUGUCAACACA----AGUUGACAA ----------........(((((.--.--....)))))........((((....))))..........((..(((...)))..))..((((((((...----.)))))))) ( -15.60, z-score = -0.60, R) >droAna3.scaffold_12916 7068511 78 + 16180835 -----------------------------CUCUGGCCAUUCCUAAUGCGUUUUAAUGCGAUAAUAAAUCAUUGGAAAUCCAUUUGUAUUGUCAACACA----AGUUGACAA -----------------------------...(((...((((....((((....))))(((.....)))...))))..)))......((((((((...----.)))))))) ( -17.30, z-score = -2.21, R) >dp4.chr4_group5 1771220 109 - 2436548 CAUUUGUCUAUCGUCUAUCGUCCA--UAUCUAUGUGCGAUUCUAAUGCGUUUUAAUGCGAUAAUAAAUCAUUGGAAAUCCAUUUGUAUUGUCAACACACAAGAGUUGACAG .((((((.((((....((((..((--(....)))..))))......((((....))))))))))))))...(((....))).......(((((((........))))))). ( -23.30, z-score = -1.59, R) >droPer1.super_5 6180740 109 - 6813705 CAUUUGUCUAUCGUCUAUCGUCCA--UAUCUAUGUGCGAUUCUAAUGCGUUUUAAUGCGAUAAUAAAUCAUUGGAAAUCCAUUUGUAUUGUCAACACACAAGAGUUGACAG .((((((.((((....((((..((--(....)))..))))......((((....))))))))))))))...(((....))).......(((((((........))))))). ( -23.30, z-score = -1.59, R) >droWil1.scaffold_180708 8431839 92 + 12563649 ---------------UCUUGUCGAGUUGUAUCAGAUCUUUCCUAAUGCGUUUUAAUGCAAUAAUAAAUCAUUGGAAAUCCAUUUGUAUUGUCAACACA----AGUUGACAA ---------------...((((((.((((................(((((....)))))((((((......(((....)))....))))))....)))----).)))))). ( -19.40, z-score = -2.08, R) >droMoj3.scaffold_6500 3863381 96 + 32352404 -------CUGCUGUUGCUCCUCU-----UGGCAGUCUUUGCCUAAUGCGUUCUAAUGCAACAAUAAAUCAUUGGAAAUCCAUUUAUAUUGUCAACACA---UGCCUGACAA -------.((.(((((.......-----.(((((...)))))...(((((....)))))((((((......(((....)))....)))))))))))))---.......... ( -16.70, z-score = -0.17, R) >droGri2.scaffold_15126 6921422 94 - 8399593 ---------GCAGCUACUGGCUG-----CUGUGCUGUUUACCUAAUGCGUUCUAAUGCAAUAAUAAAUCAUUGGAAAUCCAUUUAUAUUGUCAACACA---AGCCCGACAA ---------(((((.....))))-----)((((.((.......((((..((((((((...........))))))))...))))........)).))))---.......... ( -20.06, z-score = -0.97, R) >consensus _______CU_CUCUCACUUGUCUA__C__CCCUGGGCAUUUCUAAUGCGUUUUAAUGCUAUAAUAAAUCAUUGGAAAUCCAUUUGUAUUGUCAACACA____AGUUGACAA ...........................................((((..((((((((...........))))))))...))))....((((((((........)))))))) (-10.88 = -10.89 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:44 2011