| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,681,457 – 19,681,561 |
| Length | 104 |
| Max. P | 0.599725 |

| Location | 19,681,457 – 19,681,561 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.30 |
| Shannon entropy | 0.55622 |
| G+C content | 0.35690 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -10.72 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
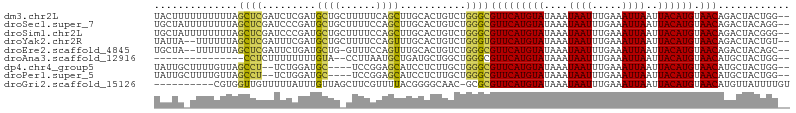
>dm3.chr2L 19681457 104 - 23011544 UACUUUUUUUUUUAGCUCGAUCUCGAUGCUGCUUUUUCAGCUUGCACUGUCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAGACUACUGG-- .............((((((....))).)))((((...(((......)))...))))(((((((((....((((.....))))..)))))).)))..........-- ( -18.60, z-score = -0.76, R) >droSec1.super_7 3296202 104 - 3727775 UGCUAUUUUUUUUAGCUCGAUCCCGAUGCUGCUUUUCCAGCUUGCACUGUCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAGACUACAGG-- .((((.......))))(((....)))(((.(((.....)))..)))..(((((......((((((....((((.....))))..))))))...)))))......-- ( -19.70, z-score = -0.32, R) >droSim1.chr2L 19349299 104 - 22036055 UGCUAUUUUUUUUAGCUCGAUCCCGAUGCUGCUUUUCCAGCUUGCACUGUCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAGACUACGGG-- .((((.......)))).....((((.(((.(((.....)))..)))..(((((......((((((....((((.....))))..))))))...)))))..))))-- ( -24.90, z-score = -1.93, R) >droYak2.chr2R 6155109 102 - 21139217 UAUUA--UUUUUUAGCUCGAUUUCGAUGCUGCUUUUCCAGUUUGCACUGUCUGGGUGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAGACUACUGU-- .....--.....(((((((....))).))))......((((.......(((((......((((((....((((.....))))..))))))...))))).)))).-- ( -18.80, z-score = -0.74, R) >droEre2.scaffold_4845 17957868 101 + 22589142 UGCUA--UUUUUUAGCUCGAUUCUGAUGCUG-GUUUCCAGUUUGCACUGUCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAGACUACAGC-- .(((.--.......(((((((..((..((((-(...)))))...))..))).))))(((((((((....((((.....))))..)))))).))).......)))-- ( -23.10, z-score = -1.32, R) >droAna3.scaffold_12916 7065940 87 + 16180835 ---------------CCUCUUUUUUUUGUA--CCUUAAUGCUGAUGCUGGCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAUGCUACUGG-- ---------------............(((--(((....(((......))).))).(((((((((....((((.....))))..)))))).))).)))......-- ( -11.70, z-score = 0.58, R) >dp4.chr4_group5 1766490 98 - 2436548 UAUUGCUUUUGUUAGCCU--UCUGGAUGC----UCCGGAGCAUCCUCUUGCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAUGCUACUGG-- ....((...(((((((((--...((((((----(....))))))).......))))...((((((....((((.....))))..))))))))))).))......-- ( -25.70, z-score = -2.32, R) >droPer1.super_5 6175532 98 - 6813705 UAUUGCUUUUGUUAGCCU--UCUGGAUGC----UCCGGAGCAUCCUCUUGCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAUGCUACUGG-- ....((...(((((((((--...((((((----(....))))))).......))))...((((((....((((.....))))..))))))))))).))......-- ( -25.70, z-score = -2.32, R) >droGri2.scaffold_15126 6917244 95 - 8399593 ----------CGUGGUUGUUUUUAUUUGUUAGCUUCGUUUUACGGGGCAAC-GCGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAUGUUAUUUUGU ----------.................(((.((((((.....)))))))))-(((.(((((((((....((((.....))))..)))))).))).)))........ ( -16.80, z-score = 0.12, R) >consensus UACUA_UUUUUUUAGCUCGAUCUCGAUGCUG_UUUUCCAGCUUGCACUGUCUGGGCGUUCAUGUAUAAAUAAUUUGAAAUUAAUUACAUGUAACAGACUACUGG__ ..............((((.........((((......))))...........))))(((((((((....((((.....))))..)))))).)))............ (-10.72 = -10.73 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:41 2011