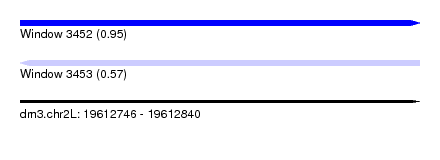
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,612,746 – 19,612,840 |
| Length | 94 |
| Max. P | 0.949311 |

| Location | 19,612,746 – 19,612,840 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.38 |
| Shannon entropy | 0.37384 |
| G+C content | 0.44674 |
| Mean single sequence MFE | -26.71 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949311 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
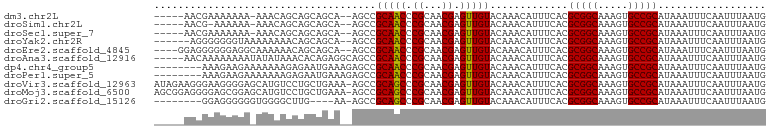
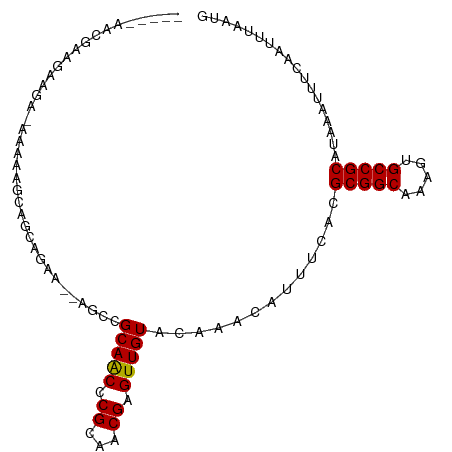
>dm3.chr2L 19612746 94 + 23011544 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU--UGCUGCUGCUGUUU-UUUUUUUCGUU----- .........((((((((((((((.....))))))))))..........((((((...))))))(((((.--....))))).....-....))))...----- ( -28.30, z-score = -2.00, R) >droSim1.chr2L 19300477 93 + 22036055 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU--UGCUGCUGCUGUUU-UUUUUU-CGUU----- .........((((((((((((((.....))))))))))..........((((((...))))))(((((.--....))))).....-...)))-)...----- ( -28.30, z-score = -1.96, R) >droSec1.super_7 3250154 94 + 3727775 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU--UGCUGCUGCUGUUU-UUUUUUUCGUU----- .........((((((((((((((.....))))))))))..........((((((...))))))(((((.--....))))).....-....))))...----- ( -28.30, z-score = -2.00, R) >droYak2.chr2R 6109698 94 + 21139217 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU--UGCUGCUGUUUUUUUUACCCCCCCU------ ...((((..(((.((((((((((.....)))))))))).........(((((((...)))))))((((.--....)))))))..))))........------ ( -26.00, z-score = -1.58, R) >droEre2.scaffold_4845 17912352 96 - 22589142 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU--UGCUGCUGUUUUUUGCCUCCCCCCUCC---- .............((((((((((.....)))))))))).(((....)))....(..(((((..(((((.--....)))))..)))))..)........---- ( -27.10, z-score = -1.78, R) >droAna3.scaffold_12916 7019290 97 - 16180835 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUGCCUCUGUGUUUAUAUAUUUUUUUUGUU----- ...(((((.((..((((((((((.....))))))))))..((..((.(((((((...)))))))..)).)).))...)))))...............----- ( -23.20, z-score = -0.81, R) >dp4.chr4_group5 1704698 94 + 2436548 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUCUUUCAUUCUCUUUUUUUCUUCUUU-------- ........(((((((((((((((.....))))))))))......((.(((((((...)))))))..))..)))))...................-------- ( -24.30, z-score = -3.00, R) >droPer1.super_5 6112501 94 + 6813705 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCUCUUUCAUUCUCUUUUUUUCUUCUUU-------- ........(((((((((((((((.....))))))))))......((.(((((((...)))))))..))..)))))...................-------- ( -24.30, z-score = -3.00, R) >droVir3.scaffold_12963 1648539 101 + 20206255 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGCUGCGGCU-UUUCAGCAGGACAUGCUCCCCUUCCCUUCUAU ........(((((((((((((((.....))))))))))...............(((((.....))))).-)))))(((.....)))................ ( -25.00, z-score = -0.19, R) >droMoj3.scaffold_6500 28562838 101 + 32352404 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGCUGCGGCU-UUUCAGCAGGACAUGCUCCGCUCCCCUCCGCU .............((((((((((.....)))))))))).(((....)))....((.(.(((..((((..-....((((.....))))))))..))).).)). ( -31.90, z-score = -1.31, R) >droGri2.scaffold_15126 1534691 89 - 8399593 CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGCUGCGGCU-UU----CAAGCCCCACCCCCCUCC-------- .............((((((((((.....)))))))))).(((....))).........(((.((.((((-..----..)))).)).))).....-------- ( -27.10, z-score = -2.20, R) >consensus CAUUAAAUUGAAAUUUAUGCGGCACUUUGCCGCGUGAAAUGUUUGUACAACUCGUUGCGGGUUGCGGCU__UGCUGCUGCUUUUU_UCUUCUUCCUU_____ .............((((((((((.....)))))))))).(((....)))....(((((.....))))).................................. (-20.20 = -20.20 + -0.00)

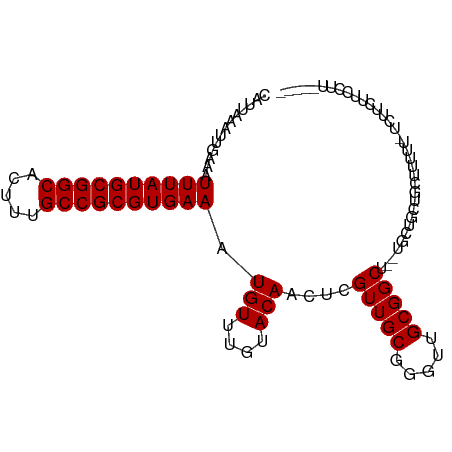
| Location | 19,612,746 – 19,612,840 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.38 |
| Shannon entropy | 0.37384 |
| G+C content | 0.44674 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.30 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565147 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 19612746 94 - 23011544 -----AACGAAAAAAA-AAACAGCAGCAGCA--AGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG -----...((((....-..(((((.((....--.)).((.....)).....))))).............(((((.....))))).....))))......... ( -19.70, z-score = -1.31, R) >droSim1.chr2L 19300477 93 - 22036055 -----AACG-AAAAAA-AAACAGCAGCAGCA--AGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG -----...(-(((...-..(((((.((....--.)).((.....)).....))))).............(((((.....))))).....))))......... ( -19.70, z-score = -1.22, R) >droSec1.super_7 3250154 94 - 3727775 -----AACGAAAAAAA-AAACAGCAGCAGCA--AGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG -----...((((....-..(((((.((....--.)).((.....)).....))))).............(((((.....))))).....))))......... ( -19.70, z-score = -1.31, R) >droYak2.chr2R 6109698 94 - 21139217 ------AGGGGGGGUAAAAAAAACAGCAGCA--AGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG ------.(.(((.((.......)).((.(..--..).))..))).)...(((.(((....))).)))..(((((.....))))).................. ( -20.80, z-score = -0.12, R) >droEre2.scaffold_4845 17912352 96 + 22589142 ----GGAGGGGGGAGGCAAAAAACAGCAGCA--AGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG ----...(((..(.(((..............--.))).)..))).....(((.(((....))).)))..(((((.....))))).................. ( -24.66, z-score = -0.97, R) >droAna3.scaffold_12916 7019290 97 + 16180835 -----AACAAAAAAAAUAUAUAAACACAGAGGCAGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG -----.......................((((.....(((((.((...)).))))).............(((((.....))))).....))))......... ( -16.40, z-score = -0.86, R) >dp4.chr4_group5 1704698 94 - 2436548 --------AAAGAAGAAAAAAAGAGAAUGAAAGAGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG --------..............((((.(((((.....(((((.((...)).))))).......))))).(((((.....))))).....))))......... ( -19.60, z-score = -2.27, R) >droPer1.super_5 6112501 94 - 6813705 --------AAAGAAGAAAAAAAGAGAAUGAAAGAGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG --------..............((((.(((((.....(((((.((...)).))))).......))))).(((((.....))))).....))))......... ( -19.60, z-score = -2.27, R) >droVir3.scaffold_12963 1648539 101 - 20206255 AUAGAAGGGAAGGGGAGCAUGUCCUGCUGAAA-AGCCGCAGCCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG .((((..(((((.((.((.(((...(((....-))).))))))).)...(((.(((....))).)))..(((((.....))))).....))))..))))... ( -27.40, z-score = -0.78, R) >droMoj3.scaffold_6500 28562838 101 - 32352404 AGCGGAGGGGAGCGGAGCAUGUCCUGCUGAAA-AGCCGCAGCCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG ...((((.(..((((.((.(((...(((....-))).)))))))))..)(((.(((....))).)))..(((((.....))))).....))))......... ( -30.50, z-score = -0.61, R) >droGri2.scaffold_15126 1534691 89 + 8399593 --------GGAGGGGGGUGGGGCUUG----AA-AGCCGCAGCCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG --------(..(.(((.((.((((..----..-)))).)).))).)..)(((.(((....))).)))..(((((.....))))).................. ( -31.80, z-score = -2.45, R) >consensus _____AACGAAGAAGA_AAAAAGCAGCAGAA__AGCCGCAACCCGCAACGAGUUGUACAAACAUUUCACGCGGCAAAGUGCCGCAUAAAUUUCAAUUUAAUG .....................................(((((.((...)).))))).............(((((.....))))).................. (-15.50 = -15.30 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:35 2011