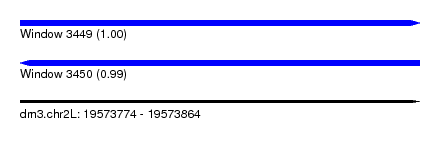
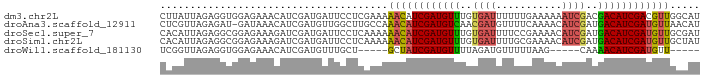
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,573,774 – 19,573,864 |
| Length | 90 |
| Max. P | 0.999352 |

| Location | 19,573,774 – 19,573,864 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.48806 |
| G+C content | 0.38440 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -14.48 |
| Energy contribution | -16.16 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
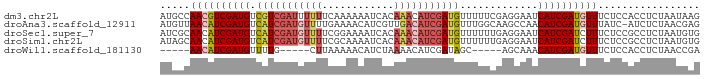
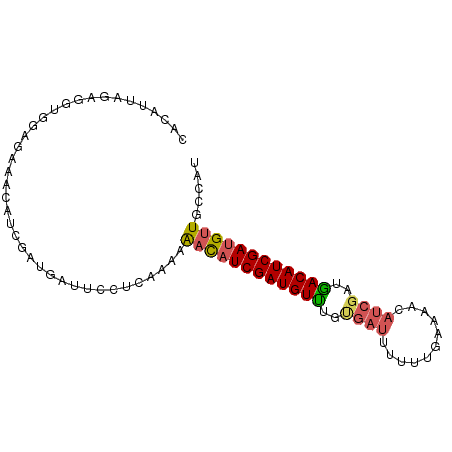
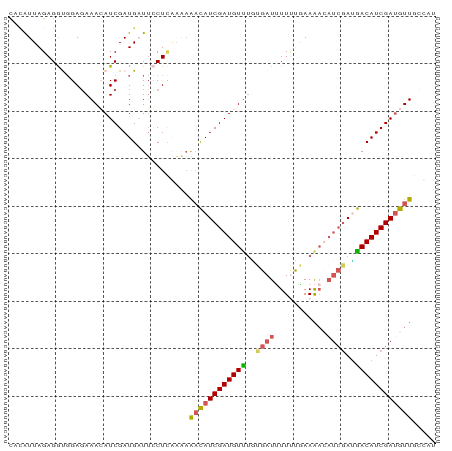
>dm3.chr2L 19573774 90 + 23011544 AUGCCAACGUCGAUGUCGUCGAUUUUUUCAAAAAAUCACAAACAUCGAUGUUUUUCGAGGAAUCAUCGAUGUUUCUCCACCUCUAAUAAG .....(((((((((((.((.((((((.....))))))))..)))))))))))....(((((((.......)))))))............. ( -21.50, z-score = -2.68, R) >droAna3.scaffold_12911 2449433 89 + 5364042 AUGUUAACAUCGAUGUCAUCGAUGUUUUGAAAACAUCGUUGACAUCGAUGUUUGGCAAGCCAACAUCGAUGUUUAUC-AUCUCUAACGAG (((..(((((((((((((.(((((((.....))))))).)))))))((((((.((....))))))))))))))...)-))(((....))) ( -37.90, z-score = -6.63, R) >droSec1.super_7 3188050 90 + 3727775 AUCGCAACAUCGAUGUCAUCGAUGUUUUCGGAAAAUCACAAACAUCGAUGUUUUUUGAGGAAUCAUCGAUCUUUCUCCGCCUCUAAUGUG ..((((...((((...(((((((((((...(.....)..)))))))))))....))))(((..............)))........)))) ( -19.24, z-score = -0.51, R) >droSim1.chr2L 19261859 90 + 22036055 AUAGCAACAUCGAUGUCAUCGAUGUUUUCGCAAAAUCACAAACAUCGAUGUUUUUUGAGGAAUCAUCGAUCUUUCUCCGCCUCUAAUGUG ...(((((((((((((....(((...........)))....)))))))))))....((((((.........)))))).)).......... ( -18.80, z-score = -1.12, R) >droWil1.scaffold_181130 7972083 75 - 16660200 -----AACAUCGAUGUUUUG-----CUUAAAAACAUCUAAAACAUCGAUAGC-----AGCAAACAUCGAUGUUUCUCCACCUCUAACCGA -----(((((((((((((((-----((......................)))-----)..)))))))))))))................. ( -17.55, z-score = -4.78, R) >consensus AUGGCAACAUCGAUGUCAUCGAUGUUUUCAAAAAAUCACAAACAUCGAUGUUUUUCGAGGAAUCAUCGAUGUUUCUCCACCUCUAAUGAG .....((((((((((.(((((((((((............))))))))))).............))))))))))................. (-14.48 = -16.16 + 1.68)
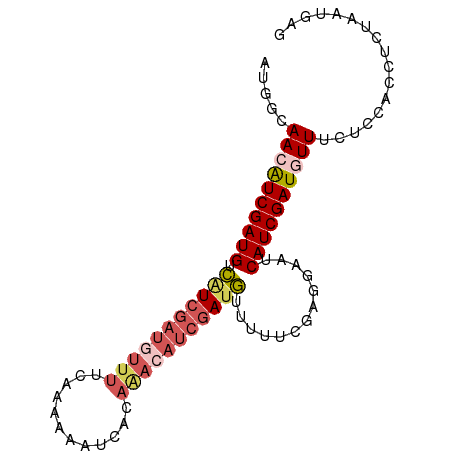
| Location | 19,573,774 – 19,573,864 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.48806 |
| G+C content | 0.38440 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -14.20 |
| Energy contribution | -14.68 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19573774 90 - 23011544 CUUAUUAGAGGUGGAGAAACAUCGAUGAUUCCUCGAAAAACAUCGAUGUUUGUGAUUUUUUGAAAAAAUCGACGACAUCGACGUUGGCAU ...((((..((((......))))..)))).....(...(((.((((((((..(((((((.....)))))))..)))))))).)))..).. ( -22.30, z-score = -1.10, R) >droAna3.scaffold_12911 2449433 89 - 5364042 CUCGUUAGAGAU-GAUAAACAUCGAUGUUGGCUUGCCAAACAUCGAUGUCAACGAUGUUUUCAAAACAUCGAUGACAUCGAUGUUAACAU (((....)))..-....((((((((((((((....)).))))))(((((((.(((((((.....))))))).)))))))))))))..... ( -36.90, z-score = -6.26, R) >droSec1.super_7 3188050 90 - 3727775 CACAUUAGAGGCGGAGAAAGAUCGAUGAUUCCUCAAAAAACAUCGAUGUUUGUGAUUUUCCGAAAACAUCGAUGACAUCGAUGUUGCGAU .(((((.((((((..((....))..))...))))......(((((((((((.((......)).))))))))))).....)))))...... ( -22.70, z-score = -0.96, R) >droSim1.chr2L 19261859 90 - 22036055 CACAUUAGAGGCGGAGAAAGAUCGAUGAUUCCUCAAAAAACAUCGAUGUUUGUGAUUUUGCGAAAACAUCGAUGACAUCGAUGUUGCUAU .......((((((..((....))..))...))))....((((((((((((..((((.((.....)).))))..))))))))))))..... ( -23.30, z-score = -1.54, R) >droWil1.scaffold_181130 7972083 75 + 16660200 UCGGUUAGAGGUGGAGAAACAUCGAUGUUUGCU-----GCUAUCGAUGUUUUAGAUGUUUUUAAG-----CAAAACAUCGAUGUU----- ...............(((((((((((((.....-----)).))))))))))).((((((((....-----.))))))))......----- ( -20.20, z-score = -2.23, R) >consensus CACAUUAGAGGUGGAGAAACAUCGAUGAUUCCUCAAAAAACAUCGAUGUUUGUGAUUUUUUGAAAACAUCGAUGACAUCGAUGUUGCCAU ......................................((((((((((((..((((.((.....)).))))..))))))))))))..... (-14.20 = -14.68 + 0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:33 2011