| Sequence ID | dm3.chr2L |
|---|---|
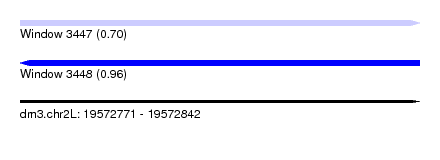
| Location | 19,572,771 – 19,572,842 |
| Length | 71 |
| Max. P | 0.957208 |

| Location | 19,572,771 – 19,572,842 |
|---|---|
| Length | 71 |
| Sequences | 4 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Shannon entropy | 0.30800 |
| G+C content | 0.39025 |
| Mean single sequence MFE | -18.23 |
| Consensus MFE | -13.98 |
| Energy contribution | -14.97 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699921 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
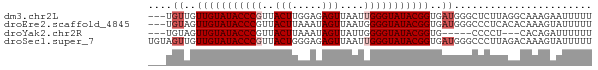
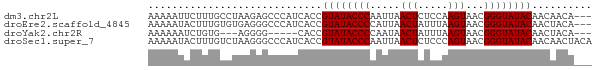
>dm3.chr2L 19572771 71 + 23011544 ---UGUUGUUGUAUACCCGUUACUUGGAGAGUUAAUUGGGUAUACGGUGAUGGGCUCUUAGGCAAAGAAUUUUU ---.((..(((((((((((..((((...))))....)))))))))))..))..(((....)))........... ( -20.50, z-score = -2.56, R) >droEre2.scaffold_4845 17872086 71 - 22589142 ---UGUAGUUGUAUACCCGUUACUUAAAUAGUUAAUGGGGUAUACGGUGAUGGGCCCUCACACAAAGUAUUUUU ---(((...(((((((((((((((.....)).)))).)))))))))((((.(....)))))))).......... ( -18.00, z-score = -0.88, R) >droYak2.chr2R 6069756 63 + 21139217 ---UGUAGUUGUAUACCCGUUACUUAAAUAGUUAUUGGGGUAUACGGUG-----CCCCU---CACAGAUUUUUU ---.(((.((((((((((...(((.....))).....))))))))))))-----)....---............ ( -14.70, z-score = -0.95, R) >droSec1.super_7 3187090 74 + 3727775 UGUAGUUGUUGUAUACCCGUUACUGGGAGAGUUAAUUGGGUAUACGGUGAUGGGCCCUUAGACAAAGUAUUUUU .((.((..(((((((((((..(((.....)))....)))))))))))..))..))................... ( -19.70, z-score = -1.34, R) >consensus ___UGUAGUUGUAUACCCGUUACUUAAAGAGUUAAUGGGGUAUACGGUGAUGGGCCCUUAGACAAAGAAUUUUU ....((..(((((((((((..(((.....)))....)))))))))))..))....................... (-13.98 = -14.97 + 1.00)
| Location | 19,572,771 – 19,572,842 |
|---|---|
| Length | 71 |
| Sequences | 4 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Shannon entropy | 0.30800 |
| G+C content | 0.39025 |
| Mean single sequence MFE | -14.32 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957208 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
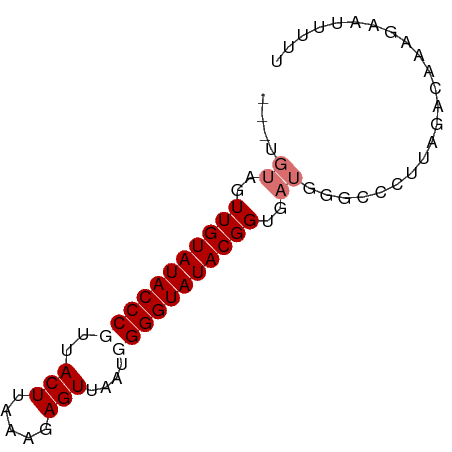
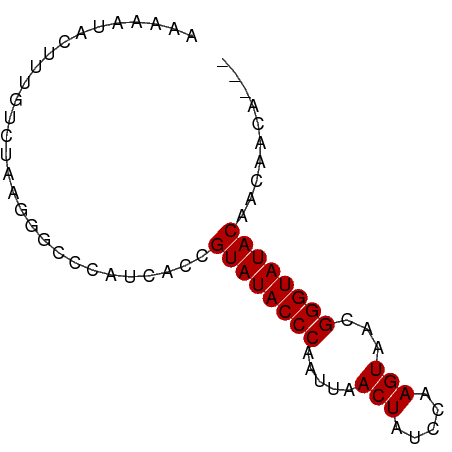
>dm3.chr2L 19572771 71 - 23011544 AAAAAUUCUUUGCCUAAGAGCCCAUCACCGUAUACCCAAUUAACUCUCCAAGUAACGGGUAUACAACAACA--- .....(((((.....))))).........((((((((.....(((.....)))...)))))))).......--- ( -13.10, z-score = -3.22, R) >droEre2.scaffold_4845 17872086 71 + 22589142 AAAAAUACUUUGUGUGAGGGCCCAUCACCGUAUACCCCAUUAACUAUUUAAGUAACGGGUAUACAACUACA--- ...........(.(((((....).)))))((((((((.....(((.....)))...)))))))).......--- ( -15.40, z-score = -1.05, R) >droYak2.chr2R 6069756 63 - 21139217 AAAAAAUCUGUG---AGGGG-----CACCGUAUACCCCAAUAACUAUUUAAGUAACGGGUAUACAACUACA--- .........(((---.((..-----..))((((((((.....(((.....)))...))))))))...))).--- ( -14.60, z-score = -1.42, R) >droSec1.super_7 3187090 74 - 3727775 AAAAAUACUUUGUCUAAGGGCCCAUCACCGUAUACCCAAUUAACUCUCCCAGUAACGGGUAUACAACAACUACA .........((((....((........))((((((((.....(((.....)))...)))))))).))))..... ( -14.20, z-score = -1.83, R) >consensus AAAAAUACUUUGUCUAAGGGCCCAUCACCGUAUACCCAAUUAACUAUCCAAGUAACGGGUAUACAACAACA___ .................((........))((((((((.....(((.....)))...)))))))).......... (-12.00 = -12.25 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:31 2011