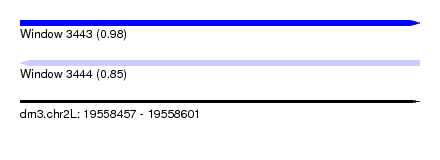
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,558,457 – 19,558,601 |
| Length | 144 |
| Max. P | 0.978132 |

| Location | 19,558,457 – 19,558,601 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 151 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Shannon entropy | 0.27842 |
| G+C content | 0.46015 |
| Mean single sequence MFE | -49.38 |
| Consensus MFE | -39.32 |
| Energy contribution | -40.40 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
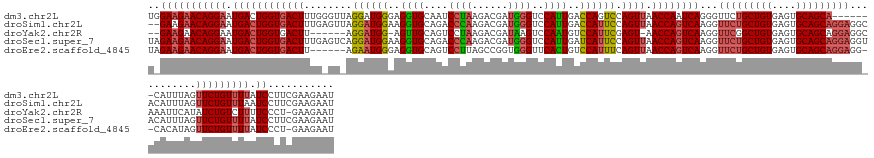
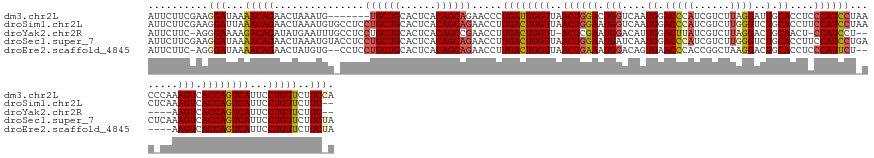
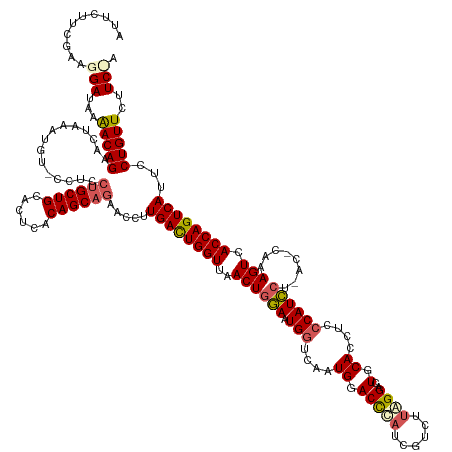
>dm3.chr2L 19558457 144 + 23011544 UGGAAGAACAGGAAUGACUGGUGACUUUGGGUUAGGAUGGGAGGUGCAAUCCUAAGACGAUGGGUCCAUUGACCAGUCCAGUUAACCAAUCAGGGUUCUGCUGUGAGUGCAGCA-------CAUUUAGUUCUGUUUUAUCCUUCGAAGAAU .((((((((((((.(((.((..((((((((((((((((.(((((......)))....(((((....))))).)).))))...)))))...))))))).((((((....))))))-------)).))).))))))))).))).......... ( -44.20, z-score = -0.79, R) >droSim1.chr2L 19246035 149 + 22036055 --GAAGAACAGGAAUGACUGGUGACUUUGAGUUAGGAUGGAAGGUGCAGACCCAAGACGAUGGGUCCAUUGACCAUUCCAGUUAACCAGUCAAGGUUCUGCUGUGAGUGCAGCAGGAGGCACAUUUAGUUCUGUUUAAUCCUUCGAAGAAU --(((((((((((.(((((((((((.....))).((((((..((((..((((((......))))))))))..))))))......))))))))...(((((((((....)))))))))...........))))))).....))))....... ( -52.80, z-score = -3.12, R) >droYak2.chr2R 6054647 140 + 21139217 --GAAGAACAGGAAUGACUGGUGACUU------AGGAUGG-AGUUGCAGUCCUAAGACGAUAAGUCCAAUGUCCAUUCGAGU-AACCAGUCAAGGUUCGGCUGUGAGUGCAGCAGGAGGCAAAUUCAUAUCUGUCUUUUCCCU-GAAGAAU --......((((..((((((((.((((------.((((((-(((((..(((....)))((....)))))).)))))))))))-.)))))))).......(((((....)))))((((((((.((....)).)))))))).)))-)...... ( -45.40, z-score = -2.23, R) >droSec1.super_7 3172515 151 + 3727775 UAGAAGAACAGGAAUGACUGGUGACUUUGAGUCAGGAUGGAAGGUGCAGACCCAAGACGAUGGGUCCAUUGAUCAUUCCAGUUAACCAGUCAAGGUUCUGCUGUGAGUGCAGCAGGAGGUACAUUUAGUUCUGUUUUAUCCUUCGAAGAAU ..(((((((((((.(((((((((((.....))).((((((..((((..((((((......))))))))))..))))))......))))))))...(((((((((....)))))))))...........))))))).....))))....... ( -53.00, z-score = -3.05, R) >droEre2.scaffold_4845 17856907 142 - 22589142 UAGAAGAACAGGAAUGACUGGUGACUU------AGAAUGGGAGGUGCAGUCCUUAGCCGGUGGGUUCACUGUCCAUUUCAGUUAACCAGUCAAGGUUCUGCUGUGAGUGCAGCAGGAGG--CACAUAGUUCUGUUUUAUCCCU-GAAGAAU ....(((((((((.((((((((((((.------.((((((..((((.(..((.........))..)))))..)))))).)))).))))))))...(((((((((....)))))))))..--.......)))))))))......-....... ( -51.50, z-score = -2.61, R) >consensus U_GAAGAACAGGAAUGACUGGUGACUUUG_GU_AGGAUGGAAGGUGCAGUCCUAAGACGAUGGGUCCAUUGACCAUUCCAGUUAACCAGUCAAGGUUCUGCUGUGAGUGCAGCAGGAGG_ACAUUUAGUUCUGUUUUAUCCUUCGAAGAAU ..(((((((((((.((((((((((((........((((((..((((....((((......))))..))))..)))))).)))).))))))))...(((((((((....)))))))))...........))))))))).))........... (-39.32 = -40.40 + 1.08)
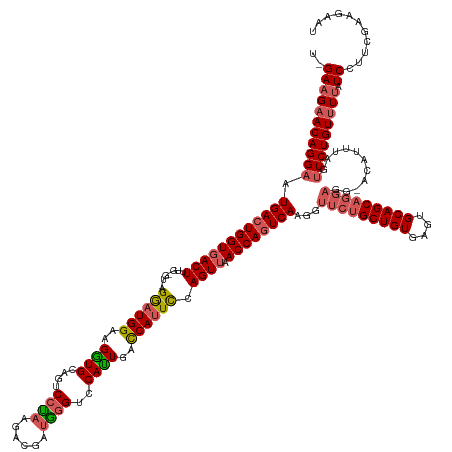
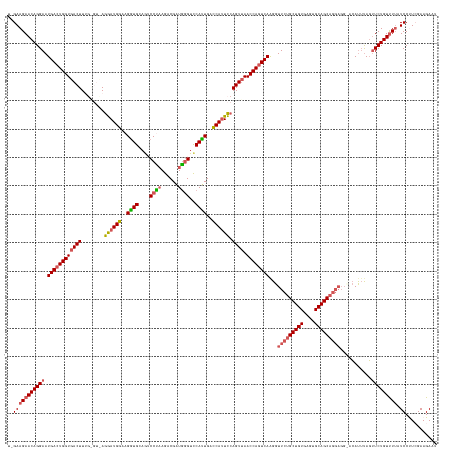
| Location | 19,558,457 – 19,558,601 |
|---|---|
| Length | 144 |
| Sequences | 5 |
| Columns | 151 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Shannon entropy | 0.27842 |
| G+C content | 0.46015 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19558457 144 - 23011544 AUUCUUCGAAGGAUAAAACAGAACUAAAUG-------UGCUGCACUCACAGCAGAACCCUGAUUGGUUAACUGGACUGGUCAAUGGACCCAUCGUCUUAGGAUUGCACCUCCCAUCCUAACCCAAAGUCACCAGUCAUUCCUGUUCUUCCA ..........(((...(((((.......(.-------(((((......))))).)....((((((((..(((.((..((((....))))..))(..(((((((.(......).)))))))..)..))).))))))))...)))))..))). ( -38.30, z-score = -1.96, R) >droSim1.chr2L 19246035 149 - 22036055 AUUCUUCGAAGGAUUAAACAGAACUAAAUGUGCCUCCUGCUGCACUCACAGCAGAACCUUGACUGGUUAACUGGAAUGGUCAAUGGACCCAUCGUCUUGGGUCUGCACCUUCCAUCCUAACUCAAAGUCACCAGUCAUUCCUGUUCUUC-- .......(((((.....((((...............((((((......)))))).....((((((((....(((((.(((...((((((((......)))))))).)))))))).....((.....)).))))))))...)))))))))-- ( -43.90, z-score = -2.94, R) >droYak2.chr2R 6054647 140 - 21139217 AUUCUUC-AGGGAAAAGACAGAUAUGAAUUUGCCUCCUGCUGCACUCACAGCCGAACCUUGACUGGUU-ACUCGAAUGGACAUUGGACUUAUCGUCUUAGGACUGCAACU-CCAUCCU------AAGUCACCAGUCAUUCCUGUUCUUC-- ......(-(((((..((.(((((....))))).)).(.((((......)))).).....((((((((.-..((.(((....))).))......(.(((((((.((.....-.))))))------))).)))))))))))))))......-- ( -35.10, z-score = -0.97, R) >droSec1.super_7 3172515 151 - 3727775 AUUCUUCGAAGGAUAAAACAGAACUAAAUGUACCUCCUGCUGCACUCACAGCAGAACCUUGACUGGUUAACUGGAAUGAUCAAUGGACCCAUCGUCUUGGGUCUGCACCUUCCAUCCUGACUCAAAGUCACCAGUCAUUCCUGUUCUUCUA .......((((((((.....................((((((......)))))).....((((((((....(((((........(((((((......))))))).....)))))....(((.....)))))))))))....)))))))).. ( -45.52, z-score = -3.60, R) >droEre2.scaffold_4845 17856907 142 + 22589142 AUUCUUC-AGGGAUAAAACAGAACUAUGUG--CCUCCUGCUGCACUCACAGCAGAACCUUGACUGGUUAACUGAAAUGGACAGUGAACCCACCGGCUAAGGACUGCACCUCCCAUUCU------AAGUCACCAGUCAUUCCUGUUCUUCUA ......(-((((((...(((......))).--....((((((......))))))......(((((((..(((..(((((...(((......((......))....)))...)))))..------.))).))))))))))))))........ ( -38.10, z-score = -1.44, R) >consensus AUUCUUCGAAGGAUAAAACAGAACUAAAUGU_CCUCCUGCUGCACUCACAGCAGAACCUUGACUGGUUAACUGGAAUGGUCAAUGGACCCAUCGUCUUAGGACUGCACCUCCCAUCCU_AC_CAAAGUCACCAGUCAUUCCUGUUCUUC_A ..........(((...(((((...............((((((......)))))).....((((((((..((((((.(((....((.(((((......))))..).))....))))))........))).))))))))...)))))..))). (-26.92 = -27.68 + 0.76)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:28 2011