| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,549,764 – 19,549,863 |
| Length | 99 |
| Max. P | 0.984835 |

| Location | 19,549,764 – 19,549,863 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 51.11 |
| Shannon entropy | 0.98176 |
| G+C content | 0.42537 |
| Mean single sequence MFE | -13.97 |
| Consensus MFE | -7.51 |
| Energy contribution | -7.53 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.984835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19549764 99 + 23011544 -----UCCCGCGCAUUAUAGCCUUAUAAUAUCUGUGCUCUUAUUU--CCCAUCCUUUUU-GCCCUCAUU---UGACCACCAAUCCAAU--CAAAAUGCGUCGCGCAGACGCU -----....(((.((((((....)))))).(((((((........--..((.......)-)........---.(((((..........--.....)).))))))))))))). ( -14.16, z-score = -1.26, R) >droGri2.scaffold_15126 7861604 112 - 8399593 AACCGCAUUUUAUAUUUGUUCACAAGUCGCUAUUCAAUUACAAGUUUUUGCUAUUUUCUCGUUUACUCCCCGUUAACUAAAACCAAAAAACAAAAUGCGACGCUUAGACGCU ...((((((((..(((((....)))))................((((((....((((...(((.((.....)).))).))))...))))))))))))))..((......)). ( -14.30, z-score = -0.67, R) >droMoj3.scaffold_6500 21328327 91 - 32352404 ----------UACCACAAUCUAUAGACAUACAUUUAACUACAAGUUAUUGCUGUUU---CGUUCAUUUC----UAUUUUAAACCAA----CAAAAUGCGUCGUUCAGGCGCU ----------...........(((((.........(((..(((....)))..))).---........))----)))..........----......(((((.....))))). ( -11.77, z-score = -0.88, R) >droWil1.scaffold_180772 5674645 93 - 8906247 -------------AAGUAAAAAUAACAAUUUGUGUUACUUUUUCUCGGUUAUCUUCGUU-CUCGCCCGC---UGCCUGUGAACUAAAA--UAAAAUGCGUCGCGCAGACGCU -------------.((.((((.(((((.....))))).)))).))...((((.((.(((-(..((....---.))....)))).)).)--)))...(((((.....))))). ( -15.00, z-score = 0.36, R) >droPer1.super_5 3556541 95 + 6813705 -------UUCAAAGUGUUUCCCCUUUUGUAAUUUUAUUUUUAUUUUGCUCCUCCUU----GUCCCCUUU---CGGCCGCCA-CACACA--CAAAAUGCGUCGCGCAGACGCC -------......((((..........((((...((....))..))))........----((..((...---.))..)).)-)))...--......(((((.....))))). ( -12.10, z-score = -0.13, R) >dp4.chr4_group1 3628731 95 - 5278887 -------UUCAAAGUGUUUCCCCUUUUGUAAUUUUAUUUUUAUUUUGCUCCUCCUU----GUCCCCUUU---CGGCCGCCA-CACACA--CAAAAUGCGUCGCGCAGACGCC -------......((((..........((((...((....))..))))........----((..((...---.))..)).)-)))...--......(((((.....))))). ( -12.10, z-score = -0.13, R) >droAna3.scaffold_12916 4152550 83 - 16180835 -------------------UCCCAGGUGUAC---UAAUUU--GGUUUUUAUUUUAUUUUCAUCGAUAUCG-AUCAUCACACGCACA----CAAAAUGCGUCGCGCAGACGCU -------------------..((((((....---..))))--))................((((....))-)).............----......(((((.....))))). ( -15.30, z-score = -0.84, R) >droEre2.scaffold_4845 17840577 99 - 22589142 -------UCCCGC-UUUUAGCCUUAUUAUAUCUGUGCUCUUAUUUUGCCGCUCCUUUCCCGCCCUCCUU---UGACCAACAACGCAAA--CGAAAUGCGUCGCGCAGACGCU -------....((-.....)).........(((((((............((.........)).......---.........(((((..--.....))))).))))))).... ( -15.60, z-score = -0.44, R) >droYak2.chr2R 6046004 85 + 21139217 -------UCCCGAAUUAUAGCCUUAUUAUAUCUGUGCUUUUAUUU--UCCCUUCUU----------------UGACCACCAACGCAUU--CGAAAUGCGUCGCGCAGACGCU -------.......................(((((((........--.........----------------((.....))((((((.--....)))))).))))))).... ( -15.40, z-score = -1.78, R) >consensus _______U_CCAAAUUUUACCCUUAUUAUAUCUGUAAUUUUAUUUUGCUCCUCCUU____GUCCACUUC___UGACCACCAACACAAA__CAAAAUGCGUCGCGCAGACGCU ................................................................................................(((((.....))))). ( -7.51 = -7.53 + 0.03)

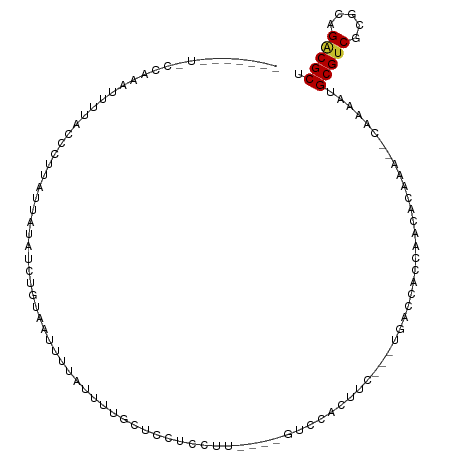
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:26 2011