
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,533,069 – 19,533,168 |
| Length | 99 |
| Max. P | 0.659038 |

| Location | 19,533,069 – 19,533,168 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.12 |
| Shannon entropy | 0.54860 |
| G+C content | 0.35828 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -12.34 |
| Energy contribution | -13.35 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.604263 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
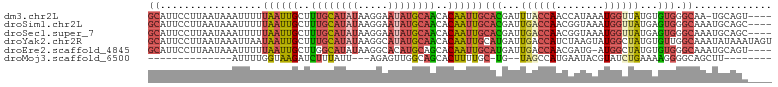
>dm3.chr2L 19533069 99 + 23011544 GCAUUCCUUAAUAAAUUUUUAAUUGCUUUGCAUAUAAGGAAUAUGCAACACAAUUGCACGAUUUACCAACCAUAAAUGGUUAUGUGUGGGCAA-UGCAGU---- (((((((............((((((..((((((((.....))))))))..))))))((((....((((........))))....)))))).))-)))...---- ( -26.80, z-score = -2.38, R) >droSim1.chr2L 19220324 100 + 22036055 GCAUUCCUUAAUAAAUUUUUAAUUGCUUUGCAUAUAAGGAAUAUGCAACACAAUUGCACGAUUGACCAACGGUAAAUGGUUAUGAGUGGGCAAAUGCAGC---- (((((((............((((((..((((((((.....))))))))..))))))(((...((((((........))))))...)))))..)))))...---- ( -26.80, z-score = -2.38, R) >droSec1.super_7 3147713 100 + 3727775 GCAUUCCUUAAUAAAUUUUUAAUUGCUUUGCAUAUAAGGAAUAUGCAACACAAUUGCACGAUUGACCAACGGUAAAUGGUUAUGAGUGGGCAAAUGCAGC---- (((((((............((((((..((((((((.....))))))))..))))))(((...((((((........))))))...)))))..)))))...---- ( -26.80, z-score = -2.38, R) >droYak2.chr2R 6029713 104 + 21139217 GCAUUCCUUAAUAAAUUAAUAAUUGCUUUGCAUAUAAGGCAUAUGCAACACAAUUGCAUGAUUGACCAUCUAAGUAUGGCUAUGUGUUGGCAAAUAUAAAUAGU ..............(((.(((.(((((..(((((((.(.(((((((((.....))))(((......)))....))))).)))))))).))))).))).)))... ( -20.70, z-score = 0.24, R) >droEre2.scaffold_4845 17823032 99 - 22589142 GCAUUCCUUAAUAAAUUUUUAAUUGCUUGGCAUAUAAGGCACAUGCAGCACAAUUGCAUGAUUGACCAACGAUG-AUGGCUAUGUGUGGGCAAAUGCAGU---- ((((...((((.......))))((((.(.(((((((.(.(((((((((.....)))))))((((.....)))).-.)).)))))))).)))))))))...---- ( -22.80, z-score = 0.79, R) >droMoj3.scaffold_6500 16191916 76 + 32352404 --------------AUUUUGGUAAGAUCUUUAUU---AGAGUUGGCAGCACUUUUGC-UG--UAGCCAUGAAUACGUAUCUGAAAAGGGGCAGCUU-------- --------------(((((((((((...))))))---)))))..((((((....)))-))--).((((((....))).(((....)))))).....-------- ( -16.40, z-score = 0.10, R) >consensus GCAUUCCUUAAUAAAUUUUUAAUUGCUUUGCAUAUAAGGAAUAUGCAACACAAUUGCACGAUUGACCAACGAUAAAUGGCUAUGAGUGGGCAAAUGCAGC____ ((.................((((((..((((((((.....))))))))..))))))(((...((((((........))))))...))).))............. (-12.34 = -13.35 + 1.01)


| Location | 19,533,069 – 19,533,168 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 71.12 |
| Shannon entropy | 0.54860 |
| G+C content | 0.35828 |
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -12.72 |
| Energy contribution | -14.41 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659038 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 19533069 99 - 23011544 ----ACUGCA-UUGCCCACACAUAACCAUUUAUGGUUGGUAAAUCGUGCAAUUGUGUUGCAUAUUCCUUAUAUGCAAAGCAAUUAAAAAUUUAUUAAGGAAUGC ----...(((-((.((......((((((....))))))((((((..(..((((((.((((((((.....)))))))).)))))).)..))))))...))))))) ( -26.00, z-score = -2.17, R) >droSim1.chr2L 19220324 100 - 22036055 ----GCUGCAUUUGCCCACUCAUAACCAUUUACCGUUGGUCAAUCGUGCAAUUGUGUUGCAUAUUCCUUAUAUGCAAAGCAAUUAAAAAUUUAUUAAGGAAUGC ----...((((((...(((.....((((........)))).....))).((((((.((((((((.....)))))))).))))))..............)))))) ( -23.20, z-score = -1.76, R) >droSec1.super_7 3147713 100 - 3727775 ----GCUGCAUUUGCCCACUCAUAACCAUUUACCGUUGGUCAAUCGUGCAAUUGUGUUGCAUAUUCCUUAUAUGCAAAGCAAUUAAAAAUUUAUUAAGGAAUGC ----...((((((...(((.....((((........)))).....))).((((((.((((((((.....)))))))).))))))..............)))))) ( -23.20, z-score = -1.76, R) >droYak2.chr2R 6029713 104 - 21139217 ACUAUUUAUAUUUGCCAACACAUAGCCAUACUUAGAUGGUCAAUCAUGCAAUUGUGUUGCAUAUGCCUUAUAUGCAAAGCAAUUAUUAAUUUAUUAAGGAAUGC ..............((........(((((......))))).........((((((.(((((((((...))))))))).)))))).............))..... ( -19.10, z-score = 0.31, R) >droEre2.scaffold_4845 17823032 99 + 22589142 ----ACUGCAUUUGCCCACACAUAGCCAU-CAUCGUUGGUCAAUCAUGCAAUUGUGCUGCAUGUGCCUUAUAUGCCAAGCAAUUAAAAAUUUAUUAAGGAAUGC ----...((((((((.........((((.-......)))).......))((((((...(((((((...)))))))...))))))..............)))))) ( -18.49, z-score = 1.26, R) >droMoj3.scaffold_6500 16191916 76 - 32352404 --------AAGCUGCCCCUUUUCAGAUACGUAUUCAUGGCUA--CA-GCAAAAGUGCUGCCAACUCU---AAUAAAGAUCUUACCAAAAU-------------- --------...............((((...((((..((((..--.(-(((....)))))))).....---))))...)))).........-------------- ( -9.90, z-score = 0.16, R) >consensus ____ACUGCAUUUGCCCACACAUAACCAUUUAUCGUUGGUCAAUCAUGCAAUUGUGUUGCAUAUUCCUUAUAUGCAAAGCAAUUAAAAAUUUAUUAAGGAAUGC .......((((((((.........((((........)))).......))((((((.((((((((.....)))))))).))))))..............)))))) (-12.72 = -14.41 + 1.69)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:25 2011