
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 175,350 – 175,453 |
| Length | 103 |
| Max. P | 0.993772 |

| Location | 175,350 – 175,446 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Shannon entropy | 0.57229 |
| G+C content | 0.43322 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.62 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.918272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
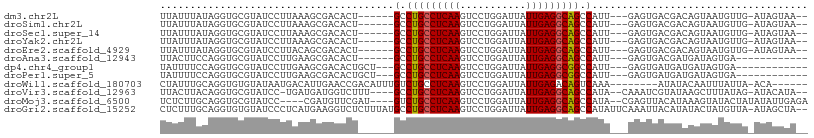
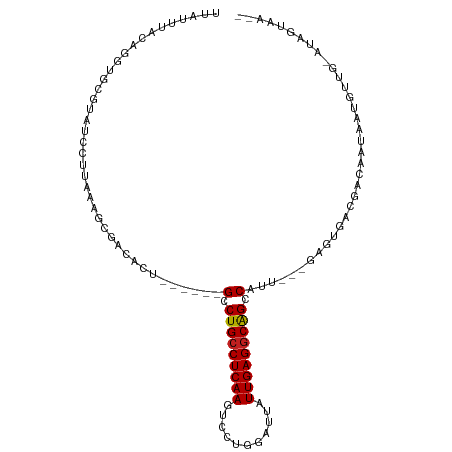
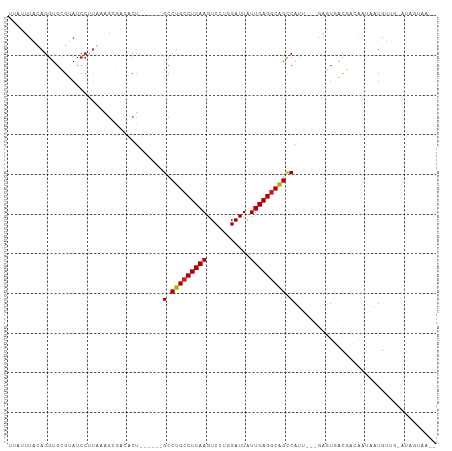
>dm3.chr2L 175350 96 + 23011544 UUAUUUAUAGGUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU---GAGUGACGACAGUAAUGUUG-AUAGUAA-- (((((((..(((((((.........))).))))------(.(((((((((...........))))))))).)..)---))))))(((((....)))))-.......-- ( -27.60, z-score = -2.08, R) >droSim1.chr2L 178739 96 + 22036055 UUAUUUAUAGGUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU---GAGUGACGACAGUAAUGUUG-AUAGUAA-- (((((((..(((((((.........))).))))------(.(((((((((...........))))))))).)..)---))))))(((((....)))))-.......-- ( -27.60, z-score = -2.08, R) >droSec1.super_14 174160 96 + 2068291 UUAUUUAUAGGUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU---GAGUGACGACAGUAAUGUUG-AUAGUAA-- (((((((..(((((((.........))).))))------(.(((((((((...........))))))))).)..)---))))))(((((....)))))-.......-- ( -27.60, z-score = -2.08, R) >droYak2.chr2L 165697 96 + 22324452 UUAUUUAUAGGUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU---GAGUGACGACAGUAAUGUUG-AUAGUAA-- (((((((..(((((((.........))).))))------(.(((((((((...........))))))))).)..)---))))))(((((....)))))-.......-- ( -27.60, z-score = -2.08, R) >droEre2.scaffold_4929 219535 96 + 26641161 UUAUUUAUAGGUGCGUAUCCUUACAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU---GAGUGACGACAGUAAUGUUG-AUAGUAA-- (((((((..(((((((.........))).))))------(.(((((((((...........))))))))).)..)---))))))(((((....)))))-.......-- ( -27.60, z-score = -1.80, R) >droAna3.scaffold_12943 811416 87 - 5039921 UUACUUCCAGGUGCGUAUCCUUGAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU---GAGUGACGAUGAUAGUGA------------ ...((((.(((.......))).))))...((((------(.(((((((((...........))))))))).((((---(.....))))).))))).------------ ( -27.30, z-score = -1.92, R) >dp4.chr4_group1 818361 90 - 5278887 UAUUUUCCAGGUGCGUAUCCUUGAAGCGACACUGCU---GCCUGCCUCAAGUCCUGGAUUAUUGAGGCGGCCAUU---GAGUGAUGAUGAUAGUGA------------ ...((((.(((.......))).))))...(((((..---(((.(((((((...........))))))))))((((---....))))....))))).------------ ( -24.50, z-score = -0.41, R) >droPer1.super_5 749877 90 + 6813705 UAUUUUCCAGGUGCGUAUCCUUGAAGCGACACUGCU---GCCUGCCUCAAGUCCUGGAUUAUUGAGGCGGCCAUU---GAGUGAUGAUGAUAGUGA------------ ...((((.(((.......))).))))...(((((..---(((.(((((((...........))))))))))((((---....))))....))))).------------ ( -24.50, z-score = -0.41, R) >droWil1.scaffold_180703 2291066 93 + 3946847 CUAUUUGCAGGUGUGUAUAAUGACAUUGAACCGACAUUUGUCUGCCUCAAGUCCUGGAUUAUUGAGACAGUCAAA--------AUAUACAAUUUAUUA-ACA------ ...........(((((((..((((........(((....)))((.(((((...........))))).))))))..--------)))))))........-...------ ( -17.20, z-score = -0.26, R) >droVir3.scaffold_12963 11263724 98 - 20206255 UUACUUACAGGUGCGUAUCC-UGAUGAUGGUCUUU----GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUA--CAAAUCGUAUAAGCUUUAUAG-AUACAUA-- ...((((((((.......))-))(((((......(----(.(((((((((...........))))))))).))..--...))))).))))........-.......-- ( -24.40, z-score = -0.90, R) >droMoj3.scaffold_6500 2390672 98 + 32352404 UCUCUUGCAGGUGCGUAUCC----CGAUGUUCGAU----GUCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUA--CGAGUUACAUAAAGUAUACUAUAUAUUGAGA ((((..((....))(((((.----..(((((((((----(.(((((((((...........))))))))).))).--)))...))))...).))))........)))) ( -27.70, z-score = -1.73, R) >droGri2.scaffold_15252 12847238 105 + 17193109 CUCUUUGCAGGUGUGUAUCCCUCAUGAAGGUCUCUUUAUGCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUAUUCAAAUUACAUAUACUAUGUUA-AUAGCUA-- ((.((.(((.(((((((..(((.....)))......((((.(((((((((...........))))))))).)))).......)))))))....))).)-).))...-- ( -25.40, z-score = -1.08, R) >consensus UUAUUUACAGGUGCGUAUCCUUAAAGCGACACU______GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUU___GAGUGACGACAAUAAUGUUG_AUAGUAA__ .......................................(.(((((((((...........))))))))).).................................... (-13.67 = -13.62 + -0.06)
| Location | 175,350 – 175,446 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Shannon entropy | 0.57229 |
| G+C content | 0.43322 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.11 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
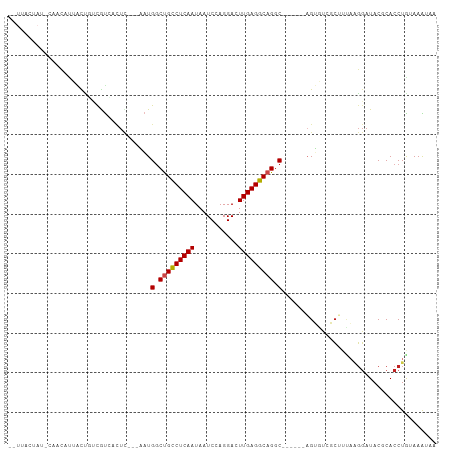
>dm3.chr2L 175350 96 - 23011544 --UUACUAU-CAACAUUACUGUCGUCACUC---AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCACCUAUAAAUAA --....(((-(....(((..(.((.((((.---...(.(((((((((...........))))))))).)------)))).)))..))).))))............... ( -23.60, z-score = -1.71, R) >droSim1.chr2L 178739 96 - 22036055 --UUACUAU-CAACAUUACUGUCGUCACUC---AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCACCUAUAAAUAA --....(((-(....(((..(.((.((((.---...(.(((((((((...........))))))))).)------)))).)))..))).))))............... ( -23.60, z-score = -1.71, R) >droSec1.super_14 174160 96 - 2068291 --UUACUAU-CAACAUUACUGUCGUCACUC---AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCACCUAUAAAUAA --....(((-(....(((..(.((.((((.---...(.(((((((((...........))))))))).)------)))).)))..))).))))............... ( -23.60, z-score = -1.71, R) >droYak2.chr2L 165697 96 - 22324452 --UUACUAU-CAACAUUACUGUCGUCACUC---AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCACCUAUAAAUAA --....(((-(....(((..(.((.((((.---...(.(((((((((...........))))))))).)------)))).)))..))).))))............... ( -23.60, z-score = -1.71, R) >droEre2.scaffold_4929 219535 96 - 26641161 --UUACUAU-CAACAUUACUGUCGUCACUC---AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUGUAAGGAUACGCACCUAUAAAUAA --....(((-(....((((.(.((.((((.---...(.(((((((((...........))))))))).)------)))).))).)))).))))............... ( -26.30, z-score = -2.10, R) >droAna3.scaffold_12943 811416 87 + 5039921 ------------UCACUAUCAUCGUCACUC---AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUCAAGGAUACGCACCUGGAAGUAA ------------...........(.((((.---...(.(((((((((...........))))))))).)------)))).)(((((.(((.......))).))))).. ( -27.10, z-score = -2.07, R) >dp4.chr4_group1 818361 90 + 5278887 ------------UCACUAUCAUCAUCACUC---AAUGGCCGCCUCAAUAAUCCAGGACUUGAGGCAGGC---AGCAGUGUCGCUUCAAGGAUACGCACCUGGAAAAUA ------------..................---.................((((((.((((((((.(((---(....))))))))))))(.....).))))))..... ( -22.90, z-score = -1.07, R) >droPer1.super_5 749877 90 - 6813705 ------------UCACUAUCAUCAUCACUC---AAUGGCCGCCUCAAUAAUCCAGGACUUGAGGCAGGC---AGCAGUGUCGCUUCAAGGAUACGCACCUGGAAAAUA ------------..................---.................((((((.((((((((.(((---(....))))))))))))(.....).))))))..... ( -22.90, z-score = -1.07, R) >droWil1.scaffold_180703 2291066 93 - 3946847 ------UGU-UAAUAAAUUGUAUAU--------UUUGACUGUCUCAAUAAUCCAGGACUUGAGGCAGACAAAUGUCGGUUCAAUGUCAUUAUACACACCUGCAAAUAG ------...-........((((((.--------..((((((((((((...........))))))))(((....)))........)))).))))))............. ( -19.20, z-score = -1.22, R) >droVir3.scaffold_12963 11263724 98 + 20206255 --UAUGUAU-CUAUAAAGCUUAUACGAUUUG--UAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC----AAAGACCAUCAUCA-GGAUACGCACCUGUAAGUAA --.......-.......(((((...(((..(--(.((.(((((((((...........))))))))).)----)...)).)))..((-((.......))))))))).. ( -24.40, z-score = -1.18, R) >droMoj3.scaffold_6500 2390672 98 - 32352404 UCUCAAUAUAUAGUAUACUUUAUGUAACUCG--UAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGAC----AUCGAACAUCG----GGAUACGCACCUGCAAGAGA ((((........((((.((..((((...(((--.(((.(((((((((...........))))))))).)----)))))))))..----))))))((....))..)))) ( -31.30, z-score = -3.64, R) >droGri2.scaffold_15252 12847238 105 - 17193109 --UAGCUAU-UAACAUAGUAUAUGUAAUUUGAAUAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGCAUAAAGAGACCUUCAUGAGGGAUACACACCUGCAAAGAG --..(((((-....)))))...((((.......((((.(((((((((...........))))))))).))))......((((....)))).........))))..... ( -26.50, z-score = -1.75, R) >consensus __UUACUAU_CAACAUUACUGUCGUCACUC___AAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC______AGUGUCGCUUUAAGGAUACGCACCUGUAAAUAA ....................................(.(((((((((...........))))))))).)....................................... (-13.02 = -13.11 + 0.09)


| Location | 175,360 – 175,453 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 71.97 |
| Shannon entropy | 0.57861 |
| G+C content | 0.43755 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -13.67 |
| Energy contribution | -13.62 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 175360 93 + 23011544 GUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU---UGAGUGACGACAGUAAUGUUGAU-AGUAAUGUAAGU-- .(((((.(((((((.(((....)------))(((((((((...........)))))))))...)---)))).))(((((....)))))..-....)))))...-- ( -26.10, z-score = -1.81, R) >droSim1.chr2L 178749 93 + 22036055 GUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU---UGAGUGACGACAGUAAUGUUGAU-AGUAAUGUAAGU-- .(((((.(((((((.(((....)------))(((((((((...........)))))))))...)---)))).))(((((....)))))..-....)))))...-- ( -26.10, z-score = -1.81, R) >droSec1.super_14 174170 93 + 2068291 GUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU---UGAGUGACGACAGUAAUGUUGAU-AGUAAUGUAAGU-- .(((((.(((((((.(((....)------))(((((((((...........)))))))))...)---)))).))(((((....)))))..-....)))))...-- ( -26.10, z-score = -1.81, R) >droYak2.chr2L 165707 93 + 22324452 GUGCGUAUCCUUAAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU---UGAGUGACGACAGUAAUGUUGAU-AGUAAUGUAAGU-- .(((((.(((((((.(((....)------))(((((((((...........)))))))))...)---)))).))(((((....)))))..-....)))))...-- ( -26.10, z-score = -1.81, R) >droEre2.scaffold_4929 219545 93 + 26641161 GUGCGUAUCCUUACAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU---UGAGUGACGACAGUAAUGUUGAU-AGUAAUGUAAGU-- .........((((((...((..(------(.(((((((((...........))))))))).)).---.......(((((....)))))..-.))..)))))).-- ( -27.90, z-score = -1.92, R) >droAna3.scaffold_12943 811426 84 - 5039921 GUGCGUAUCCUUGAAGCGACACU------GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU---UGAGUGACGAUGAUAGUGAUGUA-AGU----------- ..((.((((.(((..((..((.(------(.(((((((((...........))))))))).)).---)).))..))).))))))......-...----------- ( -22.80, z-score = -1.08, R) >dp4.chr4_group1 818371 87 - 5278887 GUGCGUAUCCUUGAAGCGACACUGCU---GCCUGCCUCAAGUCCUGGAUUAUUGAGGCGGCCAU---UGAGUGAUGAUGAUAGUGAUGUA-AGU----------- ...((((((((..(((((....))))---(((.(((((((...........))))))))))..)---..)).))).)))...........-...----------- ( -22.70, z-score = -0.44, R) >droPer1.super_5 749887 87 + 6813705 GUGCGUAUCCUUGAAGCGACACUGCU---GCCUGCCUCAAGUCCUGGAUUAUUGAGGCGGCCAU---UGAGUGAUGAUGAUAGUGAUGUA-AGU----------- ...((((((((..(((((....))))---(((.(((((((...........))))))))))..)---..)).))).)))...........-...----------- ( -22.70, z-score = -0.44, R) >droWil1.scaffold_180703 2291076 90 + 3946847 GUGUGUAUAAUGACAUUGAACCGACAUUUGUCUGCCUCAAGUCCUGGAUUAUUGAGACAGUCAA--------AAUAUACAAUUUAUUAAC-ACAUAUGU------ ((((((.((((((.((((........((((.(((.(((((...........))))).))).)))--------).....))))))))))))-))))....------ ( -19.52, z-score = -1.46, R) >droVir3.scaffold_12963 11263734 95 - 20206255 GUGCGUAUCC-UGAUGAUGGUCUUU----GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUA--CAAAUCGUAUAAGCUUUAUAGAU-ACAUAUAUAUAC-- ....((((((-(.(((((......(----(.(((((((((...........))))))))).))..--...)))))...)).......)))-))..........-- ( -22.81, z-score = -0.98, R) >droMoj3.scaffold_6500 2390682 95 + 32352404 GUGCGUAUCCC-GAUGUUCGAU-------GUCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUA--CGAGUUACAUAAAGUAUACUAUAUAUUGAGAUAAAUAU (((..((((.(-((((((((((-------(.(((((((((...........))))))))).))).--)))(((((.....))).))...)))))).))))..))) ( -26.20, z-score = -2.63, R) >droGri2.scaffold_15252 12847248 102 + 17193109 GUGUGUAUCCCUCAUGAAGGUCUCUUUAUGCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAUAUUCAAAUUACAUAUACUAUGUUAAU-AGCUAUGAAUGU-- (((((((..(((.....)))......((((.(((((((((...........))))))))).)))).......))))))).((((....))-))..........-- ( -24.70, z-score = -1.44, R) >consensus GUGCGUAUCCUUAAAGCGACACU______GCCUGCCUCAAGUCCUGGAUUAUUGAGGCAGCCAU___UGAGUGACGACAAUAAUGUUGAU_AGUAAUGUAAGU__ .............................(.(((((((((...........))))))))).)........................................... (-13.67 = -13.62 + -0.06)
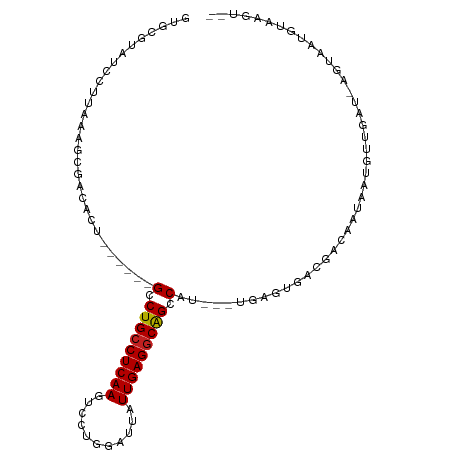
| Location | 175,360 – 175,453 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.97 |
| Shannon entropy | 0.57861 |
| G+C content | 0.43755 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.11 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 175360 93 - 23011544 --ACUUACAUUACU-AUCAACAUUACUGUCGUCACUCA---AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCAC --...........(-(((....(((..(.((.((((..---..(.(((((((((...........))))))))).)------)))).)))..))).))))..... ( -23.60, z-score = -1.92, R) >droSim1.chr2L 178749 93 - 22036055 --ACUUACAUUACU-AUCAACAUUACUGUCGUCACUCA---AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCAC --...........(-(((....(((..(.((.((((..---..(.(((((((((...........))))))))).)------)))).)))..))).))))..... ( -23.60, z-score = -1.92, R) >droSec1.super_14 174170 93 - 2068291 --ACUUACAUUACU-AUCAACAUUACUGUCGUCACUCA---AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCAC --...........(-(((....(((..(.((.((((..---..(.(((((((((...........))))))))).)------)))).)))..))).))))..... ( -23.60, z-score = -1.92, R) >droYak2.chr2L 165707 93 - 22324452 --ACUUACAUUACU-AUCAACAUUACUGUCGUCACUCA---AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUUAAGGAUACGCAC --...........(-(((....(((..(.((.((((..---..(.(((((((((...........))))))))).)------)))).)))..))).))))..... ( -23.60, z-score = -1.92, R) >droEre2.scaffold_4929 219545 93 - 26641161 --ACUUACAUUACU-AUCAACAUUACUGUCGUCACUCA---AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUGUAAGGAUACGCAC --...........(-(((....((((.(.((.((((..---..(.(((((((((...........))))))))).)------)))).))).)))).))))..... ( -26.30, z-score = -2.17, R) >droAna3.scaffold_12943 811426 84 + 5039921 -----------ACU-UACAUCACUAUCAUCGUCACUCA---AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC------AGUGUCGCUUCAAGGAUACGCAC -----------...-...(((.((.....((.((((..---..(.(((((((((...........))))))))).)------)))).)).....)))))...... ( -22.00, z-score = -1.87, R) >dp4.chr4_group1 818371 87 + 5278887 -----------ACU-UACAUCACUAUCAUCAUCACUCA---AUGGCCGCCUCAAUAAUCCAGGACUUGAGGCAGGC---AGCAGUGUCGCUUCAAGGAUACGCAC -----------...-.......................---...((((((((((...........))))))).)))---.((.(((((........))))))).. ( -21.80, z-score = -1.90, R) >droPer1.super_5 749887 87 - 6813705 -----------ACU-UACAUCACUAUCAUCAUCACUCA---AUGGCCGCCUCAAUAAUCCAGGACUUGAGGCAGGC---AGCAGUGUCGCUUCAAGGAUACGCAC -----------...-.......................---...((((((((((...........))))))).)))---.((.(((((........))))))).. ( -21.80, z-score = -1.90, R) >droWil1.scaffold_180703 2291076 90 - 3946847 ------ACAUAUGU-GUUAAUAAAUUGUAUAUU--------UUGACUGUCUCAAUAAUCCAGGACUUGAGGCAGACAAAUGUCGGUUCAAUGUCAUUAUACACAC ------.....(((-((((((..((((.((..(--------(((.(((((((((...........))))))))).)))).....)).))))...)))).))))). ( -20.30, z-score = -1.41, R) >droVir3.scaffold_12963 11263734 95 + 20206255 --GUAUAUAUAUGU-AUCUAUAAAGCUUAUACGAUUUG--UAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC----AAAGACCAUCAUCA-GGAUACGCAC --..........((-(((((((((.(......).))))--).((.(((((((((...........))))))))).)----).............-)))))).... ( -23.10, z-score = -1.46, R) >droMoj3.scaffold_6500 2390682 95 - 32352404 AUAUUUAUCUCAAUAUAUAGUAUACUUUAUGUAACUCG--UAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGAC-------AUCGAACAUC-GGGAUACGCAC .....((((((..(((((((......)))))))..(((--.(((.(((((((((...........))))))))).)-------))))).....-))))))..... ( -27.70, z-score = -3.83, R) >droGri2.scaffold_15252 12847248 102 - 17193109 --ACAUUCAUAGCU-AUUAACAUAGUAUAUGUAAUUUGAAUAUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGCAUAAAGAGACCUUCAUGAGGGAUACACAC --.........(((-((....)))))...((((.......((((.(((((((((...........))))))))).))))......((((....)))).))))... ( -26.40, z-score = -2.39, R) >consensus __ACUUACAUUACU_AUCAACAUUACUGUCGUCACUCA___AUGGCUGCCUCAAUAAUCCAGGACUUGAGGCAGGC______AGUGUCGCUUUAAGGAUACGCAC ...........................................(.(((((((((...........))))))))).)............................. (-13.02 = -13.11 + 0.09)

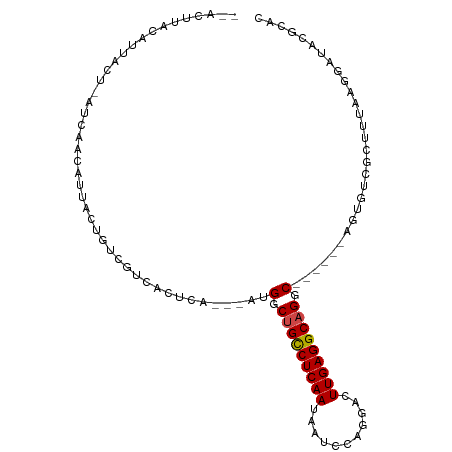
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:14 2011