
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,523,182 – 19,523,275 |
| Length | 93 |
| Max. P | 0.572937 |

| Location | 19,523,182 – 19,523,275 |
|---|---|
| Length | 93 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Shannon entropy | 0.34954 |
| G+C content | 0.46628 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -19.65 |
| Energy contribution | -19.85 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
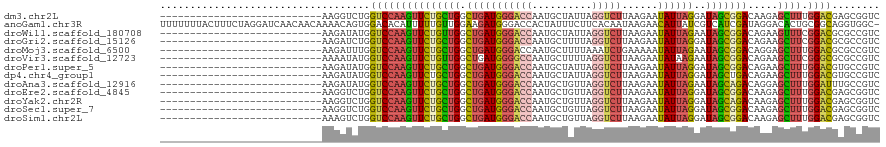
>dm3.chr2L 19523182 93 + 23011544 ---------------------------AAGGUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUAUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAAGAGCUUUGGACGAGCGGUC ---------------------------...(.((.(((((((((((.(((.(((((((........)))))))(((((((.....)))))))..)))...))))).)))))).))).... ( -31.10, z-score = -2.20, R) >anoGam1.chr3R 52787232 119 - 53272125 UUUUUUUACUUUCUAGGAUCAACAACAAAACAGUGGACACAUUUUUGUUGGAAGAUGGGACCACUAUUUCUUCACAAUAAGAACAUUAUCGUCAUCGAUAGGACACUGCGGCAGGUGGC- .....((((((.((.(......).......(((((..(...(((((((((((((((((.....)))..))))).)))))))))...(((((....))))))..))))).)).)))))).- ( -28.10, z-score = -2.57, R) >droWil1.scaffold_180708 6719497 93 + 12563649 ---------------------------AAGAUAUGGUCCAAGUUCUGUUGGCUGAUGGGACCAAUGCUAUUAGGUCUUAAGAAUAUUAGAAUAGCGGACAGAAGUUUCGGACGCGCCGUC ---------------------------........((((((.(((((((.((((.(((((((..........)))))))............)))).))))))).))..))))........ ( -25.42, z-score = -1.23, R) >droGri2.scaffold_15126 7830419 93 - 8399593 ---------------------------AAGAUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUUUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAGAAGCUUCGGACGCGCCGUC ---------------------------........(((((((((((((((.(((((((((((..........)))))......))))))..))))))).....)))).))))........ ( -25.30, z-score = 0.08, R) >droMoj3.scaffold_6500 16182491 93 + 32352404 ---------------------------AAGAUUUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUUUUAAAUCUGAAAAAUAUUAGAAUAGCGGACAGGAGCUUUGGACGCGCCGUC ---------------------------.....(((((((.((((.....))))....))))))).((.......(((((......)))))...(((..(((.....)))..))))).... ( -23.00, z-score = -0.28, R) >droVir3.scaffold_12723 2911368 93 + 5802038 ---------------------------AAAAUAUGGUCCAAGUUCUGUUGGCUGAUGGGGCCAAUGCUUUUAGGUCUUAAGAAUAUAAGAAUAGCGGACAGAAGCUUCGGGCGCGCCGUC ---------------------------........((((((((((((((.((((..(((((....)))))....(((((......))))).)))).))))).))))).))))........ ( -26.80, z-score = -0.89, R) >droPer1.super_5 3527512 93 + 6813705 ---------------------------AAGAUAUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUAUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAGAAGCUUUGGACGUGCCGUC ---------------------------........(((((((((((((((.(((((((........)))))))(((((((.....)))))))..))).)))))..)))))))........ ( -27.60, z-score = -1.49, R) >dp4.chr4_group1 3599933 93 - 5278887 ---------------------------AAGAUAUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUAUUAGGUCUUAAGAAUAUUAGGAUAGCUGACAGAAGCUUUGGACGUGCCGUC ---------------------------........((((((((((((..(((((((((........)))))..(((((((.....)))))))))))..)))))..)))))))........ ( -26.50, z-score = -1.19, R) >droAna3.scaffold_12916 4125697 93 - 16180835 ---------------------------AAGAUAUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUGUUAGGUCUUAAGAAUAUUAGAAUAGCAGACAGGAGCUUUGGAUUUGCCGUC ---------------------------..(((..((((((((((((((((.(((((((((((..........)))))......))))))..))))))).....)).)))))))....))) ( -23.40, z-score = -0.17, R) >droEre2.scaffold_4845 17813335 93 - 22589142 ---------------------------AAGGUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUGUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAAGAGCUUUGGACGAGCGGUC ---------------------------...(.((.(((((((((((.(((.(((((((........)))))))(((((((.....)))))))..)))...))))).)))))).))).... ( -30.10, z-score = -1.64, R) >droYak2.chr2R 6020032 93 + 21139217 ---------------------------AAGGUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUGUUAGGUCUUAAGAAUAUUAGGAUAGCAGACAAGAGCUUUGGACGAGCGGUC ---------------------------...(.((.(((((((((((.(((.(((((((........)))))))(((((((.....)))))))..)))...))))).)))))).))).... ( -30.80, z-score = -1.92, R) >droSec1.super_7 3137873 93 + 3727775 ---------------------------AAGGUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUGUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAAGAGCUUUGGACGAGCGGUC ---------------------------...(.((.(((((((((((.(((.(((((((........)))))))(((((((.....)))))))..)))...))))).)))))).))).... ( -30.10, z-score = -1.64, R) >droSim1.chr2L 19210374 93 + 22036055 ---------------------------AAAGUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUGUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAAGAGCUUUGGACGAGCGGUC ---------------------------...(.((.(((((((((((.(((.(((((((........)))))))(((((((.....)))))))..)))...))))).)))))).))).... ( -30.30, z-score = -1.92, R) >consensus ___________________________AAGAUCUGGUCCAAGUUCUGCUGGCUGAUGGGACCAAUGCUGUUAGGUCUUAAGAAUAUUAGGAUAGCGGACAGGAGCUUUGGACGAGCCGUC ...................................(((((((((((((((.(((((((((((..........)))))......))))))..))))))).....)))).))))........ (-19.65 = -19.85 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:24 2011