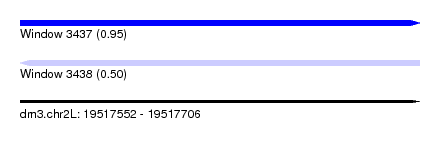
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,517,552 – 19,517,706 |
| Length | 154 |
| Max. P | 0.953381 |

| Location | 19,517,552 – 19,517,706 |
|---|---|
| Length | 154 |
| Sequences | 4 |
| Columns | 162 |
| Reading direction | forward |
| Mean pairwise identity | 64.89 |
| Shannon entropy | 0.57085 |
| G+C content | 0.36074 |
| Mean single sequence MFE | -39.16 |
| Consensus MFE | -15.71 |
| Energy contribution | -17.65 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
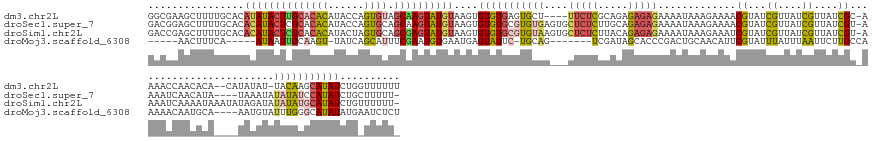
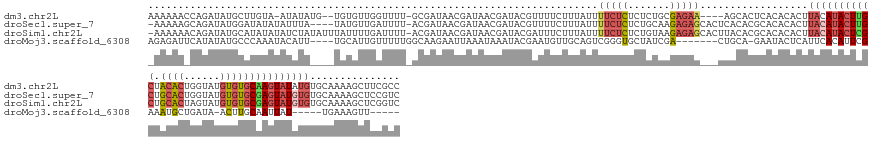
>dm3.chr2L 19517552 154 + 23011544 GGCGAAGCUUUUGCACAUAUACUUGCACACAUACCAGUGUAGCAAGUAUGUAAGUGUGUGAGUGCU----UUCUCGCAGAGAGAGAAAAUAAAGAAAACGUAUCGUUAUCGUUAUCGC-AAAACCAACACA--CAUAUAU-UACAAGCAUAUCUGGUUUUUU ...((((((..(((..((((((((((((((......)))).))))))))))..((((((...((((----(((((.....)))))).......((.((((((....)).)))).))))-)......)))))--)......-.....))).....)))))).. ( -44.40, z-score = -3.43, R) >droSec1.super_7 3132561 156 + 3727775 GACGGAGCUUUUGCACACAUACUCGCACACAUACCAGUGCAGCAAGUAUGUAAGUGUGUGCGUGUGAGUGCUCUCUUGCAGAGAGAAAAUAAAGAAAACGUAUCGUUAUCGUUAUCGU-AAAAUCAACAUA----UAAAUAUAUAUCCAUAUCUGCUUUUU- ...((((.((((((.(((.(((.(((((((((....((((.....))))....))))))))).))).)))(((((.....)))))........((.((((((....)).)))).))))-)))).)...(((----(....)))).))).............- ( -40.00, z-score = -1.63, R) >droSim1.chr2L 19201479 160 + 22036055 GACCGAGCUUUUGCACACAUACUCGCACACAUACUAGUGCAGCGAGUAUGUAAGUGUGUGCGUGUAAGUGCUCUCUUACAGAGAGAAAAUAAAGAAAUCGUAUCGUUAUCGUUAUCGU-AAAAUCAAAAUAAAUAUAGAUAUAUAUGCAUAUCUGUUUUUU- ....((((.(((((((((((((((((.(((......)))..))))))))))..((....))))))))).))))..((((.((..((..((((.((.......)).))))..)).))))-)).........(((.((((((((......)))))))).))).- ( -40.40, z-score = -1.87, R) >droMoj3.scaffold_6308 750548 139 - 3356042 -----AACUUUCA-----AUAAUUGCAAGU-UAUCAGCAUUUCGAAUGUGAAUGAGUAUUC-UGCAG-------UCGAUAGCACCCGACUGCAACAUUCGUAUUUAUUUAAUUCUUGCCAAAAACAAUGCA----AAUGUAUUUGGGCAUAUAUGAAUCUCU -----....((((-----.....(((.(((-((.........(((((((((((....))))-(((((-------(((........)))))))))))))))........)))))....(((((.(((.....----..))).))))))))....))))..... ( -31.83, z-score = -1.86, R) >consensus GACGGAGCUUUUGCACACAUACUCGCACACAUACCAGUGCAGCAAGUAUGUAAGUGUGUGCGUGCAA___CUCUCUUACAGAGAGAAAAUAAAGAAAACGUAUCGUUAUCGUUAUCGC_AAAAUCAACACA____UAUAUAUAUAUGCAUAUCUGAUUUUU_ ................((((((((((((((......)))).))))))))))....(((((((((((....(((((.....))))).............((...((....))....))........................))))))))))).......... (-15.71 = -17.65 + 1.94)
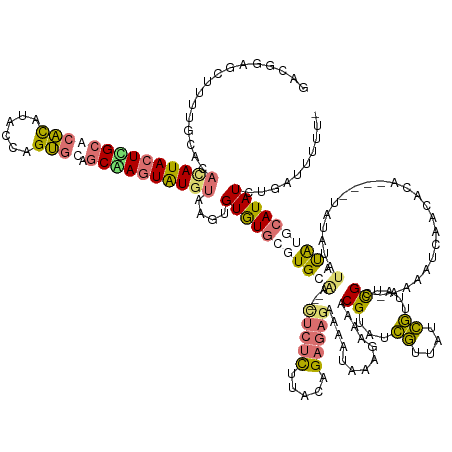
| Location | 19,517,552 – 19,517,706 |
|---|---|
| Length | 154 |
| Sequences | 4 |
| Columns | 162 |
| Reading direction | reverse |
| Mean pairwise identity | 64.89 |
| Shannon entropy | 0.57085 |
| G+C content | 0.36074 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -10.70 |
| Energy contribution | -12.83 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 19517552 154 - 23011544 AAAAAACCAGAUAUGCUUGUA-AUAUAUG--UGUGUUGGUUUU-GCGAUAACGAUAACGAUACGUUUUCUUUAUUUUCUCUCUCUGCGAGAA----AGCACUCACACACUUACAUACUUGCUACACUGGUAUGUGUGCAAGUAUAUGUGCAAAAGCUUCGCC .......((.(((((((((((-......(--(((((.(((...-(((((((.((.((((...)))).)).)))))(((((.......)))))----.))))).)))))).((((((((.........))))))))))))))))))).))............. ( -40.90, z-score = -2.44, R) >droSec1.super_7 3132561 156 - 3727775 -AAAAAGCAGAUAUGGAUAUAUAUUUA----UAUGUUGAUUUU-ACGAUAACGAUAACGAUACGUUUUCUUUAUUUUCUCUCUGCAAGAGAGCACUCACACGCACACACUUACAUACUUGCUGCACUGGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCCGUC -.....(((((.((.(((((((....)----)))))).))...-..(((((.((.((((...)))).)).))))).....)))))..(.((((........(((((((......(((((((.((((......))))))))))))))))))....)))))... ( -41.00, z-score = -1.96, R) >droSim1.chr2L 19201479 160 - 22036055 -AAAAAACAGAUAUGCAUAUAUAUCUAUAUUUAUUUUGAUUUU-ACGAUAACGAUAACGAUACGAUUUCUUUAUUUUCUCUCUGUAAGAGAGCACUUACACGCACACACUUACAUACUCGCUGCACUAGUAUGUGUGCGAGUAUGUGUGCAAAAGCUCGGUC -.......(((((((....))))))).................-..................(((...((((.....(((((.....))))).........(((((((......(((((((.((((......)))))))))))))))))).)))).)))... ( -38.10, z-score = -2.02, R) >droMoj3.scaffold_6308 750548 139 + 3356042 AGAGAUUCAUAUAUGCCCAAAUACAUU----UGCAUUGUUUUUGGCAAGAAUUAAAUAAAUACGAAUGUUGCAGUCGGGUGCUAUCGA-------CUGCA-GAAUACUCAUUCACAUUCGAAAUGCUGAUA-ACUUGCAAUUAU-----UGAAAGUU----- .....((((..(((......)))...(----((((..(((.(..(((...(((.....))).((((((((((((((((......))))-------)))))-((((....)))))))))))...)))..).)-)).)))))....-----))))....----- ( -34.40, z-score = -2.01, R) >consensus _AAAAAACAGAUAUGCAUAUAUAUAUA____UAUGUUGAUUUU_ACGAUAACGAUAACGAUACGAUUUCUUUAUUUUCUCUCUGUAAGAGAG___UUACACGCACACACUUACAUACUCGCUACACUGGUAUGUGUGCAAGUAUGUGUGCAAAAGCUCCGUC ...........................................................................(((((.......)))))..................(((((((((((.((((......)))))))))))))))............... (-10.70 = -12.83 + 2.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:23 2011