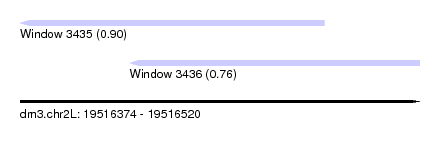
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,516,374 – 19,516,520 |
| Length | 146 |
| Max. P | 0.897874 |

| Location | 19,516,374 – 19,516,485 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 62.85 |
| Shannon entropy | 0.79484 |
| G+C content | 0.43248 |
| Mean single sequence MFE | -20.48 |
| Consensus MFE | -10.94 |
| Energy contribution | -10.37 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.46 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.897874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
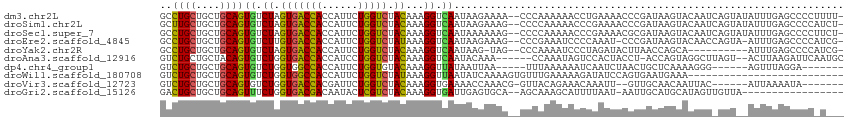
>dm3.chr2L 19516374 111 - 23011544 GCCUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAGAAAA--CCCAAAAAACCUGAAAACCCGAUAAGUACAAUCAGUAUAUUUGAGCCCCUUUU- ((((((....)))).))...(((((((......))))).))(((((((.....))...--..((((....((((.....(.....).....))))....))))....))))).- ( -17.30, z-score = 0.04, R) >droSim1.chr2L 19200387 111 - 22036055 GCUUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAGAAAG--CCCCAAAAACCCGAAAACCCGAUAAGUACAAUCAGUAUAUUUGAGCCCCAUCU- (((((.(((((...(((((.(((((((......))))).))...(((..........)--))......................)).)))..)))))....))))).......- ( -17.00, z-score = 0.31, R) >droSec1.super_7 3131464 111 - 3727775 GCCUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAAAAAG--CCCCAAAAACCCGAAAACGCGAUAAGUACAAUCAGUAUAUUUGAGCCCCUUCU- ((((((....)))).))...(((((((......))))).))...((((((........--...........((......))....((((.....))))..)))).))......- ( -17.60, z-score = 0.13, R) >droEre2.scaffold_4845 17807877 109 + 22589142 GCCUGCUGCUGCAGUGUCUUGUGACCACCAUUCUGGUCUAUAAAGGUCAAUAAGAAAG--CCCGAAAUCCCCAAAU-CCCGAUAAGUACAACCAGUA-AUUUGAGCCCCAUCG- ((((((....))))..(((((((((((((.....))).......)))).))))))..)--)...............-..((((..((.(((......-..))).))...))))- ( -18.81, z-score = -0.64, R) >droYak2.chr2R 6014699 100 - 21139217 GCCUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAG-UAG--CCCAAAAUCCCUAGAUACUUAACCAGCA----------AUUUGAGCCCCAUCG- ...(((((....(((((((((.(((((......)))))......((.(......-..)--.)).......)))))))))....)))))----------...............- ( -24.70, z-score = -2.36, R) >droAna3.scaffold_12916 4120723 105 + 16180835 GUCUGCUGCUACAGUGUCUGGUGACCACCAUCCUGGUCUACAAAGGUCAAUACAAA------CCAAAUAGUCCACUACCU-ACCAGUAGGCUUAGU--ACUUAAGAUUCAAUGC ((((((((.((.((((.((((((((((......))))).))...(((........)------))...)))..))))...)-).)))))))).....--................ ( -23.90, z-score = -0.48, R) >dp4.chr4_group1 3594995 96 + 5278887 GUCUGCUGCUGCAGUGUCUGGUGGCCACCAUUCUGGUGUACAAAGGUUAUAAUUAA-----UUUAAAAAAUCAAUCUAACUGCUCAAAAGGG------AGUUUAGGA------- ..((((....))))...((..((..((((.....))))..))..))..........-----.............(((((..((((......)------)))))))).------- ( -19.30, z-score = 0.39, R) >droWil1.scaffold_180708 6711721 88 - 12563649 GUCUGCUGCUGCAGUGUCUGGUGGCCACCAUUCUGGUCUAUAAAGGUUAAUAUCAAAAGUGUUUGAAAAAGAUAUCCAGUGAAUGAAA-------------------------- ..((((....))))...((((.(((((......)))))..............(((((....))))).........)))).........-------------------------- ( -16.00, z-score = 0.32, R) >droVir3.scaffold_12723 2906060 98 - 5802038 GUCUGCUGCUGCAGUGUCUGGUGACCACGAUUCUGGUCUACAAAGGUGAAAACCAAACG-GUUACAGAAACAAAUU--GUUGCAACAAUUAC------AUUAAAAUA------- ...((.(((.(((((.(((((((((((......))))).))...(((....))).....-....)))).....)))--)).))).)).....------.........------- ( -24.70, z-score = -1.62, R) >droGri2.scaffold_15126 7823496 94 + 8399593 GACUGCUGCUGCAGUUUCUGGUGACGACAAUACUCGUCUACAAAGGUGAUUGAGUGCA--AGCAAAGCAUUUUAAU-AAUUGCAUGCAUAGUUGUUA----------------- ((((((....)))))).....(((((((...(((((..(((....)))..)))))(((--.((((...........-..)))).)))...)))))))----------------- ( -25.52, z-score = -0.68, R) >consensus GCCUGCUGCUGCAGUGUCUGGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAGAAAG__CCCAAAAAACCCAAAUACCCGACAAGUACAAU_AGU__AUUUGAGCCCC_U___ ..((((....))))((.((.(((((((......))))).))...)).))................................................................. (-10.94 = -10.37 + -0.57)
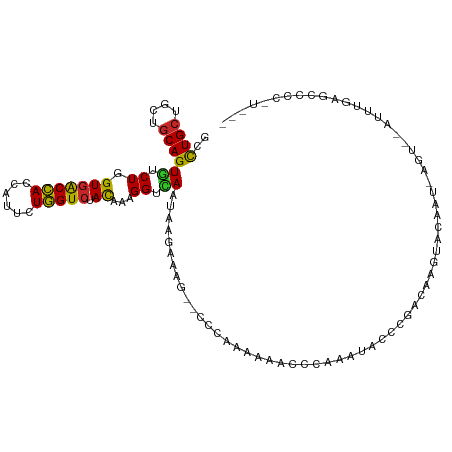

| Location | 19,516,414 – 19,516,520 |
|---|---|
| Length | 106 |
| Sequences | 10 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.08 |
| Shannon entropy | 0.64657 |
| G+C content | 0.42770 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -10.94 |
| Energy contribution | -10.37 |
| Covariance contribution | -0.57 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.53 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
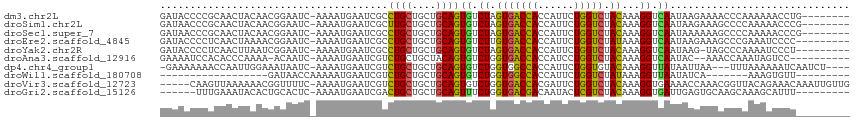
>dm3.chr2L 19516414 106 - 23011544 GAUACCCCGCAACUACAACGGAAUC-AAAAUGAAUCGCCUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAGAAAACCCAAAAAACCUG-------- (((((.(((.........)))....-.............(((....))))))))..(((((((......))))).))..((((...................)))).-------- ( -17.01, z-score = -0.16, R) >droSim1.chr2L 19200427 106 - 22036055 GAUAACCCGCAACUACAACGGAAUC-AAAAUGAAUCGCUUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAGAAAGCCCCAAAAACCCG-------- ..................(((....-.........((((.((....))))))(((..((((((((.....))).......)))))...))).............)))-------- ( -16.81, z-score = 0.26, R) >droSec1.super_7 3131504 106 - 3727775 GAUAACCCGCAACUACAACGGAAUC-AAAAUGAAUCGCCUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAAAAAGCCCCAAAAACCCG-------- (((...(((.........))).)))-....(((.((((((((....)))).))...(((((((......))))).))...)))))......................-------- ( -16.80, z-score = -0.19, R) >droEre2.scaffold_4845 17807916 105 + 22589142 GAUACCCCUCAACUAAAACGGAAUC-AAAAUGAAUCGCCUGCUGCUGCAGUGUCUUGUGACCACCAUUCUGGUCUAUAAAGGUCAAUAAGAAAGCCCGAAAUCCCC--------- ..................(((..((-.....))...((((((....))))..(((((((((((((.....))).......)))).))))))..)))))........--------- ( -20.21, z-score = -1.32, R) >droYak2.chr2R 6014730 104 - 21139217 GAUACCCCUCAACUUAAUCGGAAUC-AAAAUGAAUCGCCUGCUGCUGCAGUGUCUAGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAG-UAGCCCAAAAUCCCU--------- ...................(((.((-.....)).......((((((....((.((.(((((((......))))).))...)).))...))-))))......)))..--------- ( -18.80, z-score = -0.66, R) >droAna3.scaffold_12916 4120761 101 + 16180835 GAAAAUCCACACCCAAAA-ACAAUC-AAAAUGAAUCGUCUGCUGCUACAGUGUCUGGUGACCACCAUCCUGGUCUACAAAGGUCAAUAC--AAACCAAAUAGUCC---------- ......(((.((......-....((-.....)).......((((...)))))).))).(((((......)))))......(((......--..))).........---------- ( -11.40, z-score = 1.18, R) >dp4.chr4_group1 3595019 106 + 5278887 -GAAAAAAACCAAUUGGAAAUAAUC-AAAAUGAAUCGUCUGCUGCUGCAGUGUCUGGUGGCCACCAUUCUGGUGUACAAAGGUUAUAAUUAA---UUUAAAAAAUCAAUCU---- -......((((.((..((.....((-.....))..((.((((....))))))))..))(..((((.....))))..)...))))........---................---- ( -18.20, z-score = -0.61, R) >droWil1.scaffold_180708 6711746 81 - 12563649 ------------------GAUAACCAAAAAUGAAUCGUCUGCUGCUGCAGUGUCUGGUGGCCACCAUUCUGGUCUAUAAAGGUUAAUAUCA-------AAAGUGUU--------- ------------------..(((((........((((.((((....))))....))))(((((......)))))......)))))......-------........--------- ( -16.50, z-score = -0.67, R) >droVir3.scaffold_12723 2906079 109 - 5802038 -----CAAGUUAAAAAACGGUUUUC-AAAAUGAAUCGUCUGCUGCUGCAGUGUCUGGUGACCACGAUUCUGGUCUACAAAGGUGAAAACCAAACGGUUACAGAAACAAAUUGUUG -----.............(((((((-(....(((((((((((....)))).(((....))).)))))))((.....))....)))))))).(((((((.........))))))). ( -26.20, z-score = -1.19, R) >droGri2.scaffold_15126 7823520 99 + 8399593 ------UUUGAAAUACACUGCACUC-AAAAUGAAUCGACUGCUGCUGCAGUUUCUGGUGACGACAAUACUCGUCUACAAAGGUGAUUGAGUGCAAGCAAAGCAUUU--------- ------............(((((((-((.....(((((((((....))))))....(((((((......))))).))...)))..)))))))))............--------- ( -27.70, z-score = -1.89, R) >consensus GAUAACCCGCAACUAAAACGGAAUC_AAAAUGAAUCGCCUGCUGCUGCAGUGUCUGGUGACCACCAUUCUGGUCUACAAAGGUCAAUAAGAAAGCCCAAAAAACCC_________ ......................................((((....))))((.((.(((((((......))))).))...)).)).............................. (-10.94 = -10.37 + -0.57)
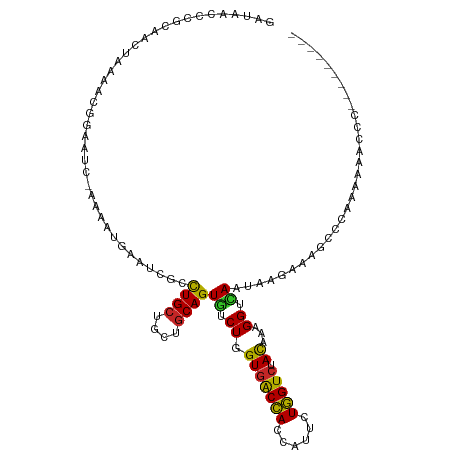
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:20 2011