
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,496,182 – 19,496,273 |
| Length | 91 |
| Max. P | 0.689469 |

| Location | 19,496,182 – 19,496,273 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Shannon entropy | 0.60777 |
| G+C content | 0.67490 |
| Mean single sequence MFE | -44.21 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.99 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
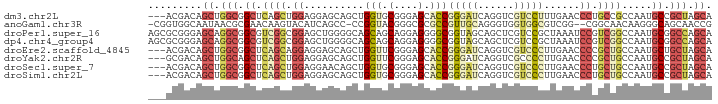
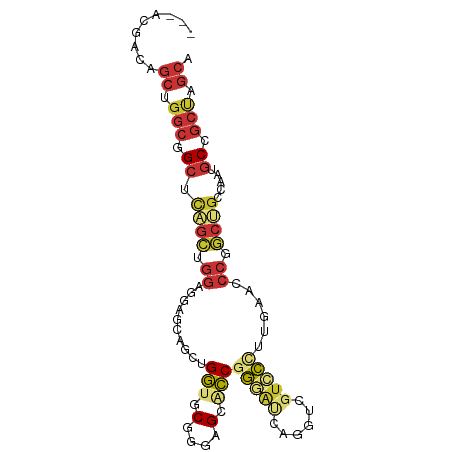
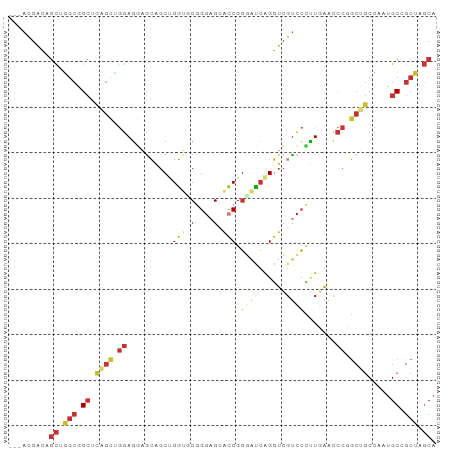
>dm3.chr2L 19496182 91 + 23011544 ---ACGACAGCUGGCGGCUCAGCUGGAGGAGCAGCUGGUGCGGGAGCACCGGGAUCAGGUCGUCCUUUGAACCCUGCCGCCAAUGCCGCUAGCA ---......(((((((((..(((((......)))))(((((....)))))(((.(((((......))))).)))..........))))))))). ( -43.70, z-score = -1.55, R) >anoGam1.chr3R 15635821 90 - 53272125 -CGGUGGCAAUAACGCGAACAAGUACAUCAGCC-CCGGUACGGGCGCGCCGUUGCAGGGUGGUGGCGUCGG--CGGCAACAAGGGCAGCAACCG -((((.((......((......))......(((-(..((.(.(.(((((((..((...))..))))).)).--).)..))..)))).)).)))) ( -32.30, z-score = 0.90, R) >droPer1.super_16 911009 94 + 1990440 AGCGCGGGAGCAGGCGGCGUCGGCGGAGCUGGGGCAGCAGCAGGAGGGGCGGUAGCAGCUCGUCCGCUAAAUCCGUCGGCCAAUGCGGCCAGCA .((((....)).(((.(((((((((((((((...))))(((.(((.((((.......)))).))))))...)))))))))....)).))).)). ( -48.80, z-score = -2.07, R) >dp4.chr4_group4 603789 94 - 6586962 AGCGCGGGAGCAGGCGGCGUCGGCGGAGCUGGGGCAGCAGCAGGAGGGGCGGUAGCAGCUCGUCCGCUAAAUCCGUCGGCCAAUGCGGCCAGCA .((((....)).(((.(((((((((((((((...))))(((.(((.((((.......)))).))))))...)))))))))....)).))).)). ( -48.80, z-score = -2.07, R) >droEre2.scaffold_4845 17788131 91 - 22589142 ---ACGACAGCUGGCGGCUCAGCAGGAGGAGCAGCUGGUUCGGGAGCACCGGGAUCAGGUCGUCCCUUGAACCCCGCUGCCAAUGCUGCUAGCA ---......(((((((((((.....)).(.(((((.(((((((.....))(((((......)))))..)))))..))))))...))))))))). ( -40.60, z-score = -0.98, R) >droYak2.chr2R 5994876 91 + 21139217 ---GCGACAGCUGGCAGCUCAGCUGGAGGAGCAGCUGGUUCGGGAGCACCGGGAUCAGGUCGCCCCUUGAACCCCGCUGCCAAUGCCGCUAGCA ---(((.((..(((((((..(((((......)))))((((((((.(((((.......))).)))))..)))))..))))))).)).)))..... ( -42.50, z-score = -1.09, R) >droSec1.super_7 3111355 91 + 3727775 ---ACGACAGCUGGCGGCUCAGCUGGAGGAACAGCUGGUGCGGGAGCACCGGGAUCAGGUCGUCCCUUGAACCCUGCUGCCAAUGCCGCUAGCA ---......((((((((((((((((......)))))))...((.((((..(((.(((((......))))).))))))).))...))))))))). ( -49.00, z-score = -3.46, R) >droSim1.chr2L 19179811 91 + 22036055 ---ACGACAGCUGGCGGCUCAGCUGGAGGAGCAGCUGGUGCGGGAGCACCGGGAUCAGGUCGUCCCUUGAACCCUGCUGCCAAUGCCGCUAGCA ---......((((((((((((((((......)))))))...((.((((..(((.(((((......))))).))))))).))...))))))))). ( -48.00, z-score = -2.60, R) >consensus ___ACGACAGCUGGCGGCUCAGCUGGAGGAGCAGCUGGUGCGGGAGCACCGGGAUCAGGUCGUCCCUUGAACCCGGCUGCCAAUGCCGCUAGCA .........((.(((.((.((((.((..........(((.(....).)))(((((......)))))......)).)))).....)).))).)). (-16.19 = -16.99 + 0.80)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:19 2011