
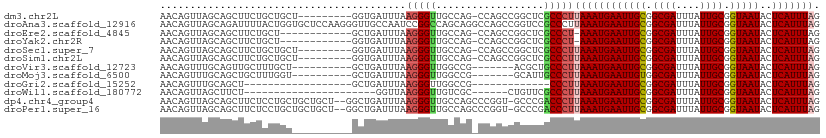
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 19,480,876 – 19,481,006 |
| Length | 130 |
| Max. P | 0.914776 |

| Location | 19,480,876 – 19,480,976 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.15 |
| Shannon entropy | 0.44278 |
| G+C content | 0.46810 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -15.85 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19480876 100 + 23011544 AACAGUUAGCAGCUUCUGCUGCU---------GGUGAUUUAAGGGUUGCCAG-CCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..((.((((((((....))))))---------))))((((((((((.(((.(-....).)))..))))))))))..(((((.((((....)))).))))).......... ( -38.00, z-score = -2.91, R) >droAna3.scaffold_12916 4082090 110 - 16180835 AACAGUUAGCAGAUUUUACUGGUGCUCCAAGGGUUGCCAAUCCGGCCAGCAGGCCAGCCGGUCCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ....................((((..((..((((.....))))((((....))))....))..))))..((((((((((((.((((....)))).)))))..))))))). ( -32.30, z-score = -0.41, R) >droEre2.scaffold_4845 17772834 96 - 22589142 AACAGUUAGCAGCUUCUGCU------------GCUGAUUUAAGGGUUGCCAG-CCAGCCGGCUCGCCCU-AAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ...((((((((((....)))------------)))))))...((((.(((.(-....).)))..))))(-(((((((((((.((((....)))).)))))..))))))). ( -37.80, z-score = -3.51, R) >droYak2.chr2R 5978945 96 + 21139217 AACAGUUAGCAGCUUCUGCU------------GGUGAUUUAAGGGUUGCCAG-CCAGCCGGCUCGCCCU-AAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ....((.(((.(((...(((------------((..(((....)))..))))-).)))..))).)).((-(((((((((((.((((....)))).)))))..)))))))) ( -34.50, z-score = -2.43, R) >droSec1.super_7 3096031 100 + 3727775 AACAGUUAGCAGCUUCUGCUGCU---------GGUGAUUUAAGGGUUGCCAG-CCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..((.((((((((....))))))---------))))((((((((((.(((.(-....).)))..))))))))))..(((((.((((....)))).))))).......... ( -38.00, z-score = -2.91, R) >droSim1.chr2L 19164500 100 + 22036055 AACAGUUAGCAGCUUCUGCUGCU---------GGUGAUUUAAGGGUUGCCAG-CCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..((.((((((((....))))))---------))))((((((((((.(((.(-....).)))..))))))))))..(((((.((((....)))).))))).......... ( -38.00, z-score = -2.91, R) >droVir3.scaffold_12723 666556 93 + 5802038 AACAGUUUGCAGUUGCUUUGCU----------GCUGAUUUAAGGGUUGGCCG-------ACGCUGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ......(((((((....(((((----------(((.((((((((((.(((..-------..))))))))))))).)...)))))))...))))))).............. ( -29.70, z-score = -2.18, R) >droMoj3.scaffold_6500 29183711 93 + 32352404 AACAGUUUGCAGCUGCUUUGGU----------GCUGAUUUAAGGGUUGGCCG-------GCAUUGCCCUUAAAUGAAUUGUGGCGAUUUAUUGCGGUAAUACUCAUUUAG .(((((((.((((.((....))----------))))((((((((((..(...-------.)...))))))))))))))))).((((....))))................ ( -25.00, z-score = -0.40, R) >droGri2.scaffold_15252 4210181 79 + 17193109 AACAGUUUGCAGCU------------------GCUGAUUUAAGGGUUGGCCG-------------CCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..((((........------------------))))((((((((((.....)-------------)))))))))..(((((.((((....)))).))))).......... ( -23.10, z-score = -1.58, R) >droWil1.scaffold_180772 7217125 82 + 8906247 AACAGUUAGCUUCU----------------------GGUUAAGGGUUGUCGC------CUGUUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..(((.......))----------------------).((((((((.(....------....).))))))))(((((((((.((((....)))).)))))..)))).... ( -19.60, z-score = -0.65, R) >dp4.chr4_group4 588921 107 - 6586962 AACAGUUAGCAGCUUCUCCUGCUGCUGCU--GGCUGAUUUAAGGGUUGCCAGCCCGGU-GCCCGACCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..(((((((((((..(....)..))))))--)))))((((((((((((...((.....-)).))))))))))))..(((((.((((....)))).))))).......... ( -42.40, z-score = -3.84, R) >droPer1.super_16 896149 107 + 1990440 AACAGUUAGCAGCUUCUCCUGCUGCUGCU--GGCUGAUUUAAGGGUUGCCAGCCCGGU-GCCCGACCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG ..(((((((((((..(....)..))))))--)))))((((((((((((...((.....-)).))))))))))))..(((((.((((....)))).))))).......... ( -42.40, z-score = -3.84, R) >consensus AACAGUUAGCAGCUUCUGCUGC__________GCUGAUUUAAGGGUUGCCAG_CCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAG .........................................(((((..................)))))((((((((((((.((((....)))).)))))..))))))). (-15.85 = -16.00 + 0.15)


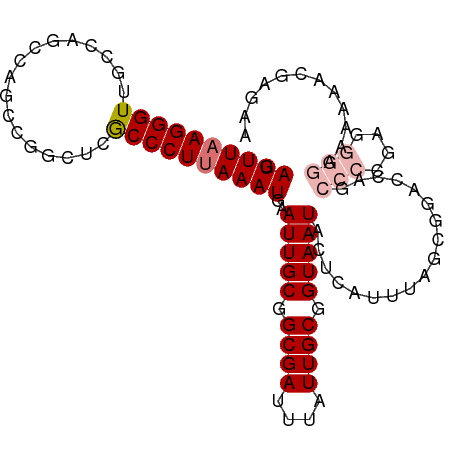
| Location | 19,480,902 – 19,481,006 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Shannon entropy | 0.39728 |
| G+C content | 0.49134 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.540620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 19480902 104 + 23011544 GAUUUAAGGGUUGCCAGCCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGCCGACCAGCCCCGAGGGGAAAAAACGAGAA .((((((((((.(((.(....).)))..))))))))))..(((((.((((....)))).))))).(((.......(.....)(((....))).......))).. ( -31.70, z-score = -0.76, R) >droEre2.scaffold_4845 17772857 103 - 22589142 GAUUUAAGGGUUGCCAGCCAGCCGGCUCGCCCU-AAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGCGGACCAGCCCCGAGGGGAAAGAACGAAUA ........((((((.((((....)))).))(((-(((((((((((.((((....)))).)))))..)))))))).)))))..(((....)))............ ( -33.30, z-score = -1.07, R) >droYak2.chr2R 5978968 103 + 21139217 GAUUUAAGGGUUGCCAGCCAGCCGGCUCGCCCU-AAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGCGGACCAGCCCCGAGGGGAAAAAACGAGAA ........(((((.....)))))..((((((((-(((((((((((.((((....)))).)))))..)))))))).)).....(((....)))......)))).. ( -33.80, z-score = -1.24, R) >droSec1.super_7 3096057 104 + 3727775 GAUUUAAGGGUUGCCAGCCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGCGGACCAGCCCCGAGGGGAAAAAACGAGAA .((((((((((.(((.(....).)))..))))))))))..(((((.((((....)))).)))))..........((......(((....)))......)).... ( -32.80, z-score = -0.90, R) >droSim1.chr2L 19164526 104 + 22036055 GAUUUAAGGGUUGCCAGCCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGCGGACCAGCCCCGAGGGGAAAAAACGAGAA .((((((((((.(((.(....).)))..))))))))))..(((((.((((....)))).)))))..........((......(((....)))......)).... ( -32.80, z-score = -0.90, R) >droGri2.scaffold_15252 4210198 88 + 17193109 GAUUUAAGGGUUGGC------------CGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGUCGCCCG----CAAGAAAAAAAAACAAAAA .((((((((((....------------.))))))))))...(((((((((((...((.(......).))...))))).)))----)))................ ( -27.60, z-score = -3.10, R) >dp4.chr4_group4 588954 80 - 6586962 GAUUUAAGGGUUGCCAGCCCGGUGCCCGACCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGUUGCCC------------------------ .((((((((((((...((.....)).)))))))))))).......(((((((...((.(......).))...))))))).------------------------ ( -24.80, z-score = -1.91, R) >droPer1.super_16 896182 80 + 1990440 GAUUUAAGGGUUGCCAGCCCGGUGCCCGACCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGUUGCCC------------------------ .((((((((((((...((.....)).)))))))))))).......(((((((...((.(......).))...))))))).------------------------ ( -24.80, z-score = -1.91, R) >consensus GAUUUAAGGGUUGCCAGCCAGCCGGCUCGCCCUUAAAUGAAUUGCGGCGAUUUAUUGCGGUAAUACUCAUUUAGCGGACCAGCCCCGAGGGGAAAAAACGAGAA .((((((((((.................))))))))))..(((((.((((....)))).)))))..................(((....)))............ (-17.94 = -18.57 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:52:18 2011